Searching NCBI's Proteins database
Go to parent Basic bioinformatics concepts, databases and tools#Exercises_during_the_training
![]() Warning: Information is liquid. Records change all the time: info is removed and added. Therefore screenshots may not be up-to-date.
Warning: Information is liquid. Records change all the time: info is removed and added. Therefore screenshots may not be up-to-date.
The Protein database is a collection of protein sequences from several sources, including translations from annotated coding regions in GenBank and RefSeq, as well as records from curated high quality protein databases like SwissProt. Since it contains translations of Genbank sequences, this means that all sequence errors in Genbank are transported to the Protein database. Fortunately, you can always check the source of the information in Protein records.
NCBI's Proteins database also contains protein sequences from the PDB database. Although PDB is a curated, high quality database, it does contain a lot of data of synthetic proteins. It's a protein structure database, containing protein annotation, sequences and 3D structures. In many cases protein structure determination is done on crystallized forms of proteins. To improve crystallization, small mutations are introduced in the proteins. As a result the protein sequences found in PDB are slight deviations from the real protein sequences as they occur in nature.
Exercise 1: heaviest human protein
| Do an advanced search to find the name of the heaviest (in terms of weight) human protein in the Protein database ? |
|---|
Note that the Molecular Weight filter was absent in the filer menu of the Nucleotide database. So the filters are automatically adjusted depending on the database that you search in. The advanced search returns all isoforms of a protein called titin, a protein of about 36000 amino acids long. |
Go to the Genbank record of the heaviest isoform.
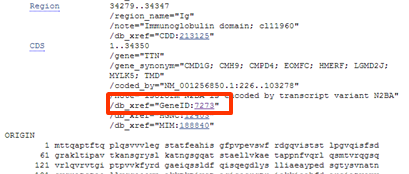
| In the CDS annotation of the FEATURES field find the cross reference to the GENE database |
|---|
| Go to the FEATURES section of the record. Look for the CDS feature (just above the sequence). There you find cross references to other databases. The second cross references to the Gene database (GeneID:7273), |
Gene is a database that contains more information on the function of the protein.
| Go to the GENE database and find the function of the protein. |
|---|
| Follow the link to go to the Gene record. In the Summary section of the Gene record you find more info on the function of the protein. |
Exercise 2: downloading multiple sequences in FASTA format
Make sure that all filters were cleared before you start the exercise.
| Search the Protein database for sugar cane (Saccharum officinarum) transcription factor sequences |
|---|
| You need an advanced search for this. You have to specify the organism and the fact that you are only interested in transcription factors. One of the ways of doing the latter is searching for "transcription factor" in the titles of the records. According to the tutorial to NCBI's Entrez system the Title field corresponds to the definition line of a record.
 This line summarizes the biology of the sequence and includes the organism, product name, gene symbol, molecule type and whether it is a partial or complete cds. So fill in the Advanced search builder as follows:
 |
This returns a set of 10 transcription factors.
| Download the sequences of the MYB transcription factors in FASTA format |
|---|
| Tick the boxes of the MYB transcription factors and expand the Send to section. Fill in the Send to section as follows:
 Click the Create File button. |