Protein ligand interaction - from small molecule to protein exercises
Exercises created by Alexander Botzki
Part of Protein Structure Analysis training
Contents
Introduction
The goal of this exercise is appreciate how protein interactions can be studied through visual inspection and other software tools. Protein interactions can be classified into different groups regarding the molecular properties and functions of the interacting partners. (These groups are intertwined in several cases.) Some examples include:
- The interactions of proteins with other proteins, small molecules, carbohydrates, lipids or nucleic acids;
- Receptor-ligand interactions;
- Antigen-antibody interactions;
- Enzymatic interactions, enzyme-inhibitor interactions.
Exploring the structure of a nanobody-stabilized active state of the β2 adrenoceptor - the ligand
We will start with exploring one crystal structure of the β2 adrenoceptor. Together with the Steyaert lab from VIB, Kobilka published several crystal structures of the β2 adrenoceptor in its various activation states (Rasmussen et al. Nature 2011, 477).
Download one crystal structure 3P0G from the PDB into Yasara.
File - Load - PDB file from internet
As you can immediately appreciate, it is a bigger crystal structure with more than one molecule.
How many molecules are present in the crystallized structures? And how many chain identifiers are used?
Some software routines need seperate chain identifiers for molecular entities to work correctly, so I suggest to rename the small molecule to chain L. You can achieve this via right click on the chain in the Head-up display and select Rename. Enter ‘L’ for instance to proceed with the renaming.
We first have a look whether we can find out if there are specific interactions of the small molecule ligand with the adrenoreceptor.
In order to do so, we first have to add Hydrogens to all present molecules.
Edit - Add – hydrogens to: All
It is preferable to view small molecules as sticks, so change the display of the ligand to Sticks.
Furthermore, we select all residues in a sphere of 10 Å around the ligand.
Select one atom of the ligand in the main display and right-click on it:
Select – in sphere around – Residue and drag with the mouse until the display says 10 Å
After having selected the ligand and the surrounding residues ( = binding pocket) display the hydrogen bonds via
View – show interactions – hydrogen bonds of - Residues
In the following dialogue select 'Selected' in the panel Belongs to or has! The selected flag option in the following window can be confirmed with OK.
Between which amino acids and the ligand do you see hydrogen bonds?
Given that hydrogen bonding is dependent on the definition of a hydrogen bond in the program, it is not a bad idea to use other tools to compare the analysis. There are many options to do this online if you look at published crystal structures. Next to the tools which are directly linked out from the web site of the crystal structure at the PDB database you can use the ProteinPlus server at http://proteinsplus.zbh.uni-hamburg.de/
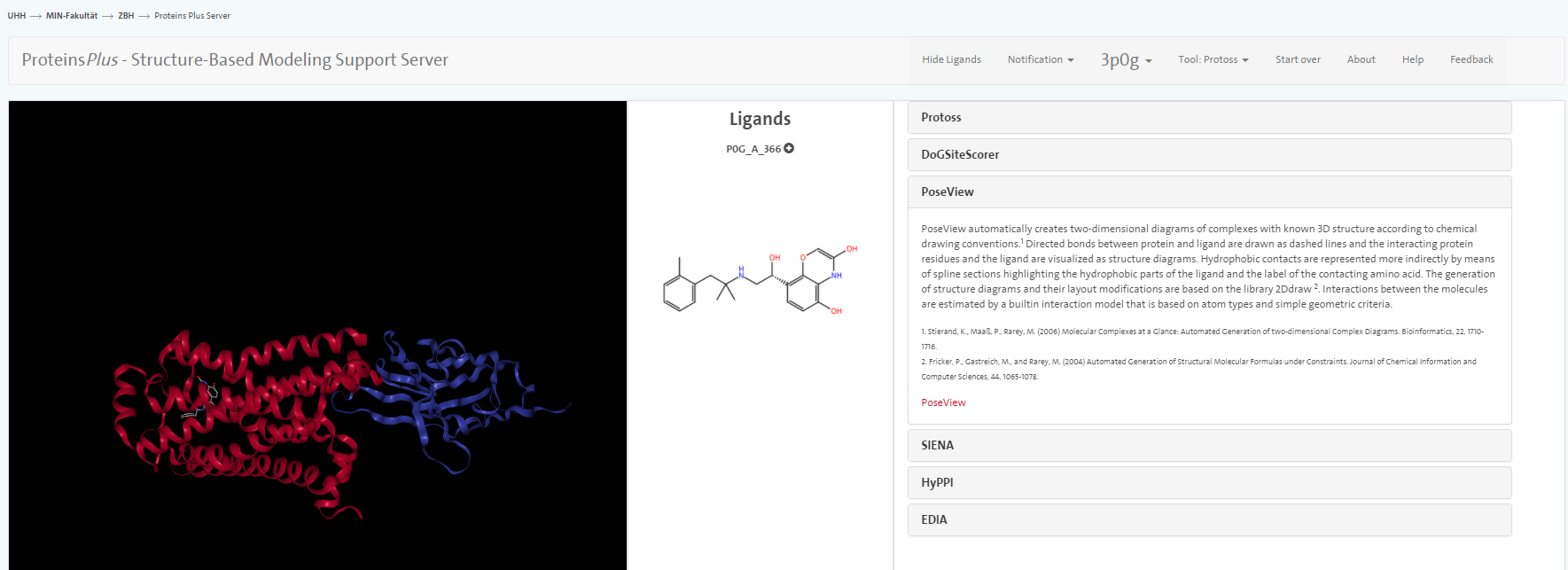
Go to the web site of ProteinPlus and enter the PDB code 3P0G into the search box. After clicking on Go, you should be presented with on overview of tools the ProteinPlus server provides.
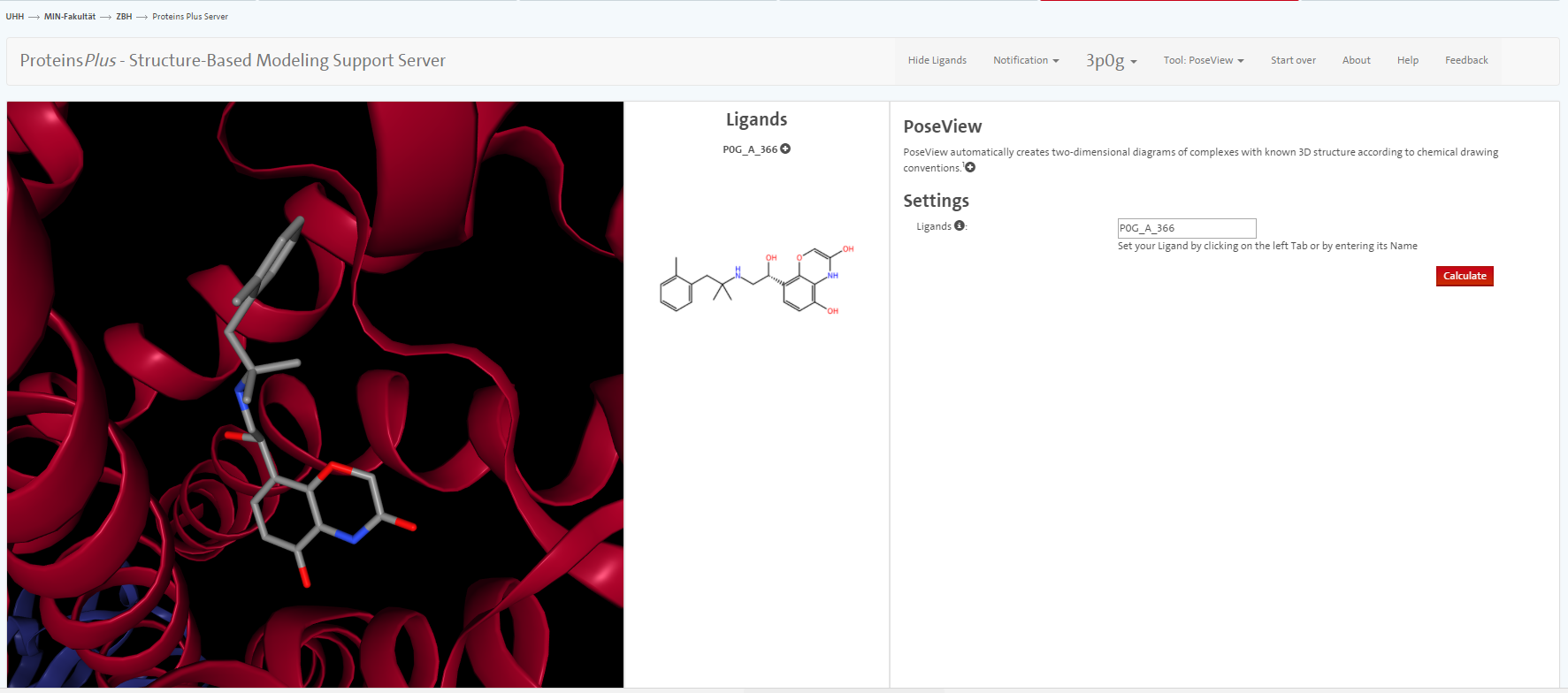
We do not go into great detail on all the tools but only mention PoseView. With this tool, you can prepare an automatic sketch of the small molecule-protein interactions.
According to PoseView, between which amino acids and the ligand do you see hydrogen bonds? What other interactions are presented in the sketch? <br> Inspect the visualisation in Yasara: Do you see the interactions in Yasara as well?
Exploring the structure of a nanobody-stabilized active state of the β2 adrenoceptor - the nanobody
In order to estimate the binding energy between the nanobody and the β2 adrenoceptor, we can use the FoldX tool AnalyseComplex. It is recommended to calculate these binding energies on energy-minimized structures. To illustrate the effect of the energy minimization, we compare the interaction energy of the current crystal structure and its minimized structure. Given that energy-minimization takes a while for this rather large complex, please download the Yasara scene here http://data.bits.vib.be/pub/trainingen/PSA/3P0G_1.sce and load it into your Yasara session.
Calculate the interaction energies between the chain A and B of the object 3P0G and the RepairObj1, respectively.
Analyze - FoldX - Interaction energy of molecules
What is the dG in the two cases? Any idea why the difference is rather hugh?
This command also creates a list of residues forming the interface of the two proteins. Hit space to see the list of residues in the interface.
Tip: This list can also be useful if you want to make visualisations of the interaction site.
Comparing the active and the inactive conformation of the β2 adrenoceptor
In case, there is still time, I would recommend to try to use some of your capabilities you learned today and create a superposition of the inactive and active conformation of the β2 adrenoceptor. We take one of crystal structures which are available: 3SN6
File - Load - PDB file from Internet
You will be kind of overwhelmed once the structure is loaded into Yasara. In order to get a first quick overview, click on the 'Center' buttom in the menu of Yasara (5th buttom from the left). Then, it is time to look at the PDB entry of 3SN6 in the PDB database to have a first idea on what molecules are in the PDB file.
As you see on the website http://www.rcsb.org/pdb/explore/explore.do?structureId=3SN6, the chain R consists of 2 molecules, the β2 adrenoceptor and lysozyme. In the corresponding article, it is stated that 'the unstructured amino terminus of the β2AR is replaced with T4 lysozyme (T4L)'.
Since this is an extra molecule in the crystal structure which disturbes our view, we will delete it.
Select the first buttom in the Yasara menu and try to select only the lysozyme.
After the manipulation, the overall picture should look roughly like this.
In the following step, we superimpose only the receptors. The rest of the structures will move along.
Analyze - Align - Pairwise, based on structure - Molecules with MUSTANG <br> First, select the first chain R from 3SN6 and second, the first chain A from 3P0G as target.
Investigate the differences in TM helices and the binding of the nanobody compared to the subunit of the G protein.
Tip: Color the secondary structures to better identify the individual chains/units of G protein.