Primer design in SnapGene tutorial
Go to parent SnapGene
Only when you have no flexibility in your choice of primers, I would use SnapGene to design them. For cloning this is often the case.
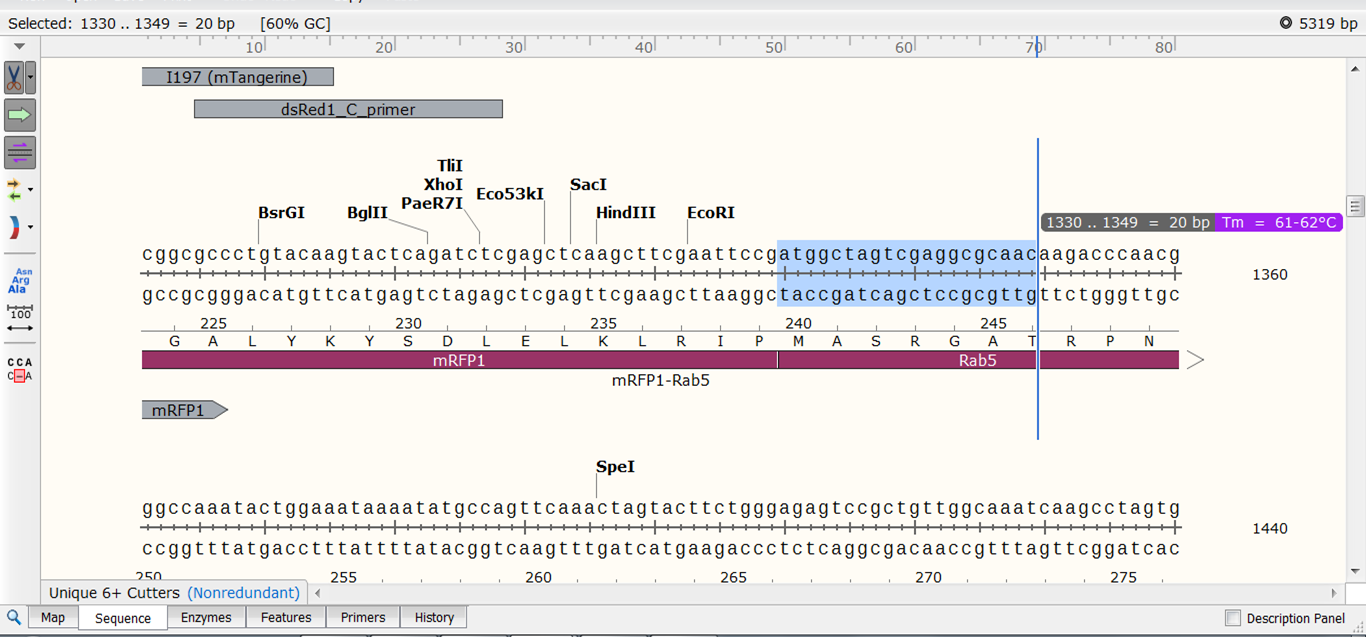
For instance, we want to make primers to clone the Rab5 gene from plasmid mRFP1-Rab5 as a fusion to the AcGFP1 gene in plasmid pAcGFP1-C1. For this, our choice of primers is restricted: the forward primer needs to start with the Rab5 start codon so its location is fixed. The forward primer should also contain a BspE1 restriction site to fuse the gene to AcGFP1. We want primers with a Tm in the range of 60-62°C.
The only thing that is still variable in this case is the length of the primer, which will be determined by the Tm that we wish.
| Design the forward primer. |
|---|
| Go back to the Sequence view of mRFP1-Rab5 and select a sequence that starts with the ATG start codon of Rab5. As you elongate your selection, SnapGene automatically displays the length and Tm of the selection:
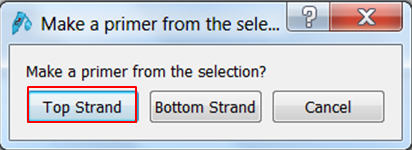
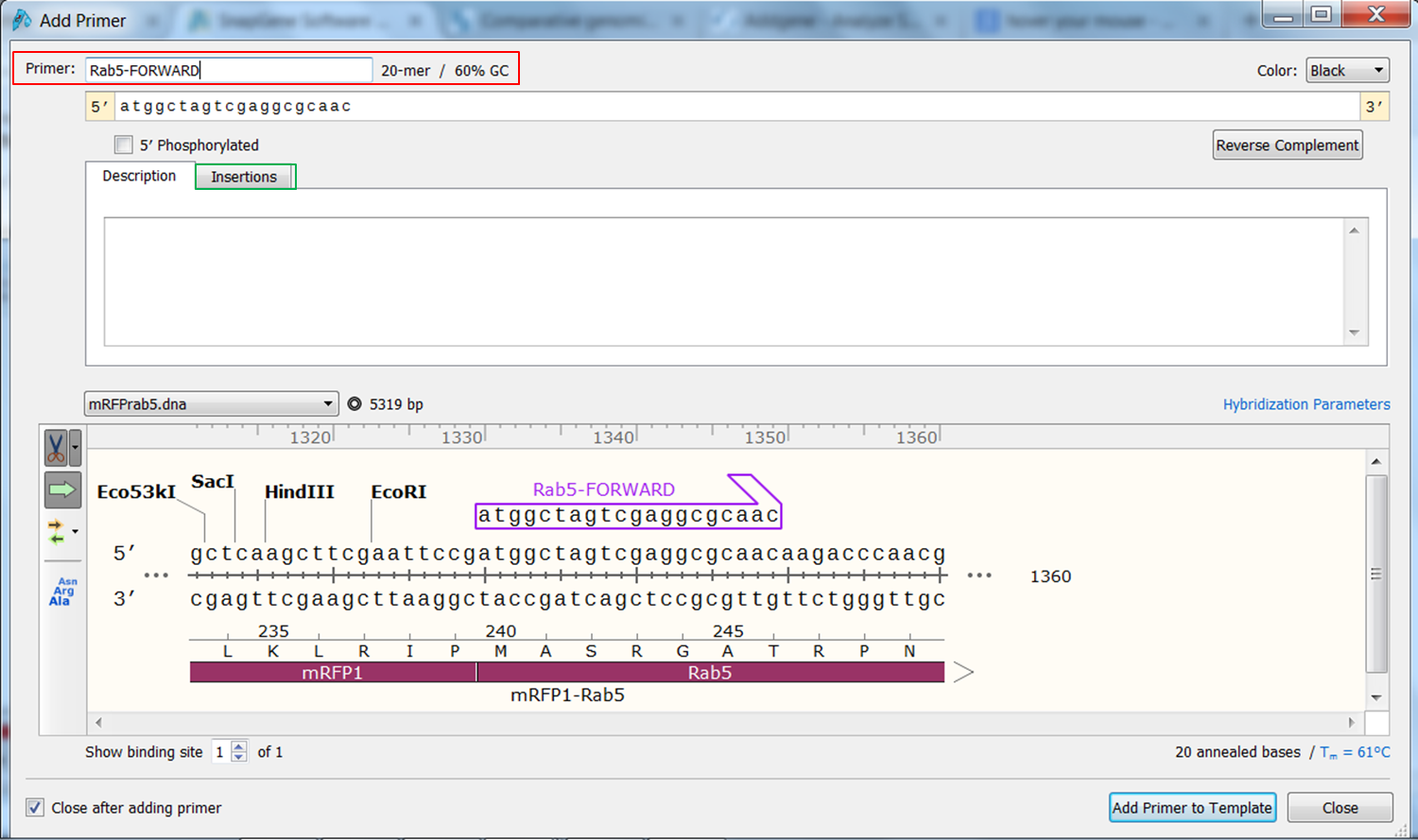
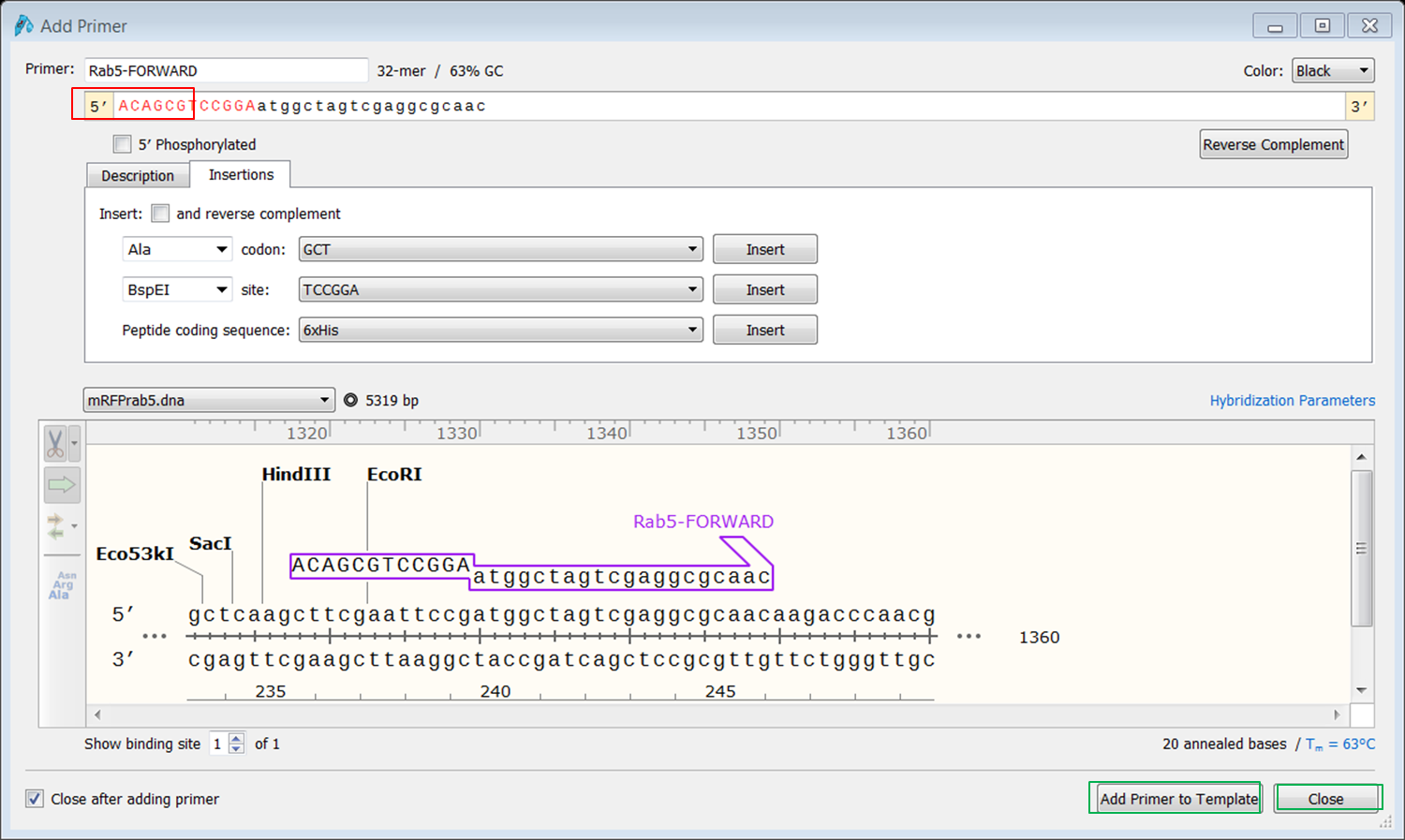
To transform the selection into a primer expand Primers in the top menu and select Add primer. Select to make a primer for the Top strand: The primer is displayed. Give it an informative name e.g. Rab5-FORWARD. To clone the gene you need a BspE1 site at the 5' end of the primer. There are two ways to add a sequence to the primer:
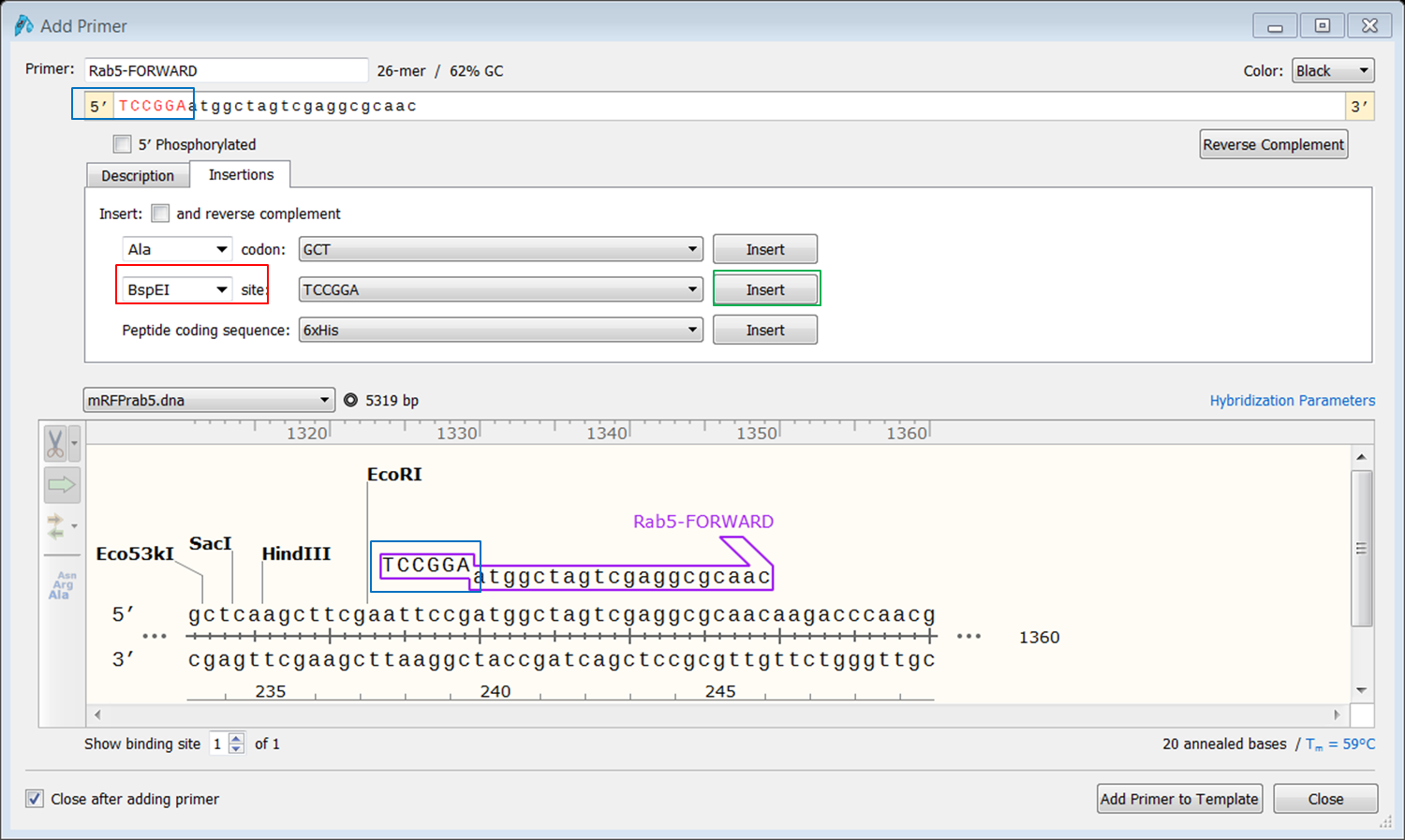
To add the restriction site select the Insertions tab: Select BspE1' in the site box (red) and click the Insert button (green): The sequence of the site is now added to the 5' end of the primer (blue). To shield the site we will some random nucleotides to the 5' end of the primer. To do this click at the 5’ end of the primer and type a random sequence of 6 nt (red): If you are happy with the sequence of the primer click Add primer to template (green). |
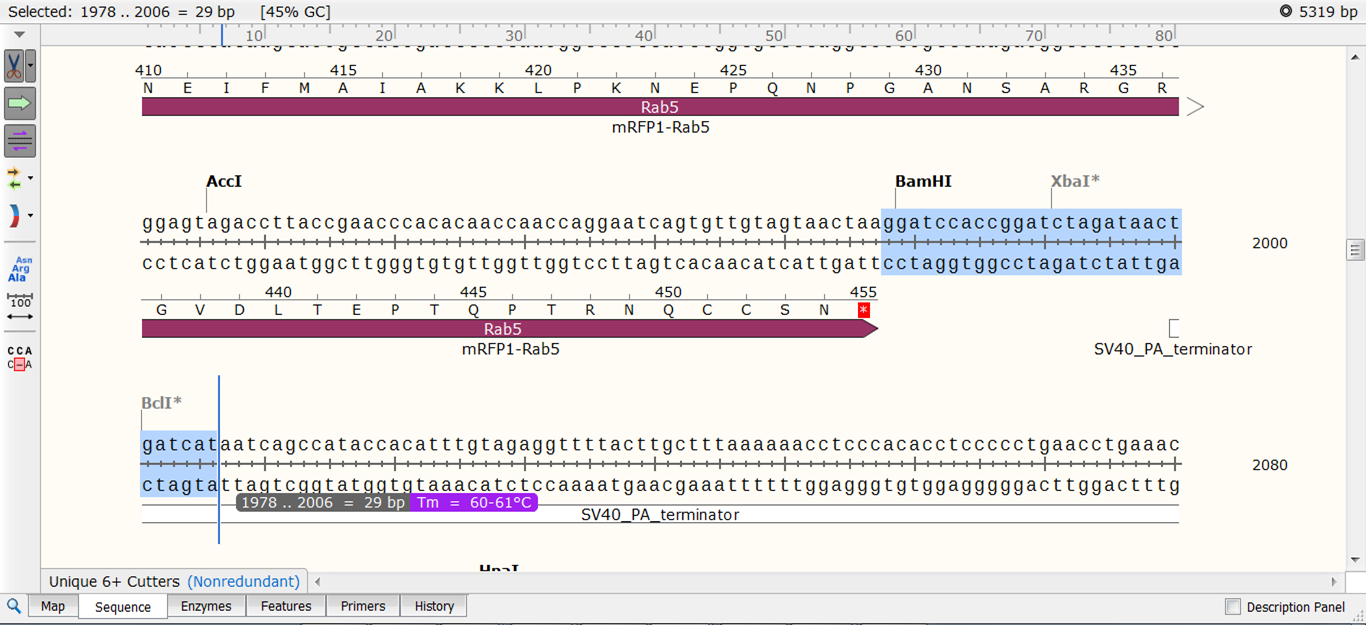
For cloning Rab5 into the vector later on, we will use a BamHI restriction site. This means we can use the existing BamH1 site right downstream of the rab5 gene. So the only requirements of the reverse primer are its location (directly after the stop codon) and its Tm (60-62°C).
Exercise: design the reverse primer
Try to do the exercise without peeking at the solution.
| Design the reverse primer. |
|---|
| Select a sequence that starts after the stop codon of Rab5 with the correct Tm:
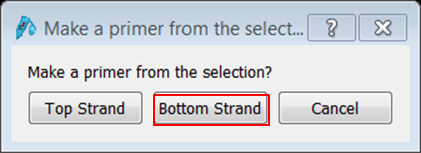
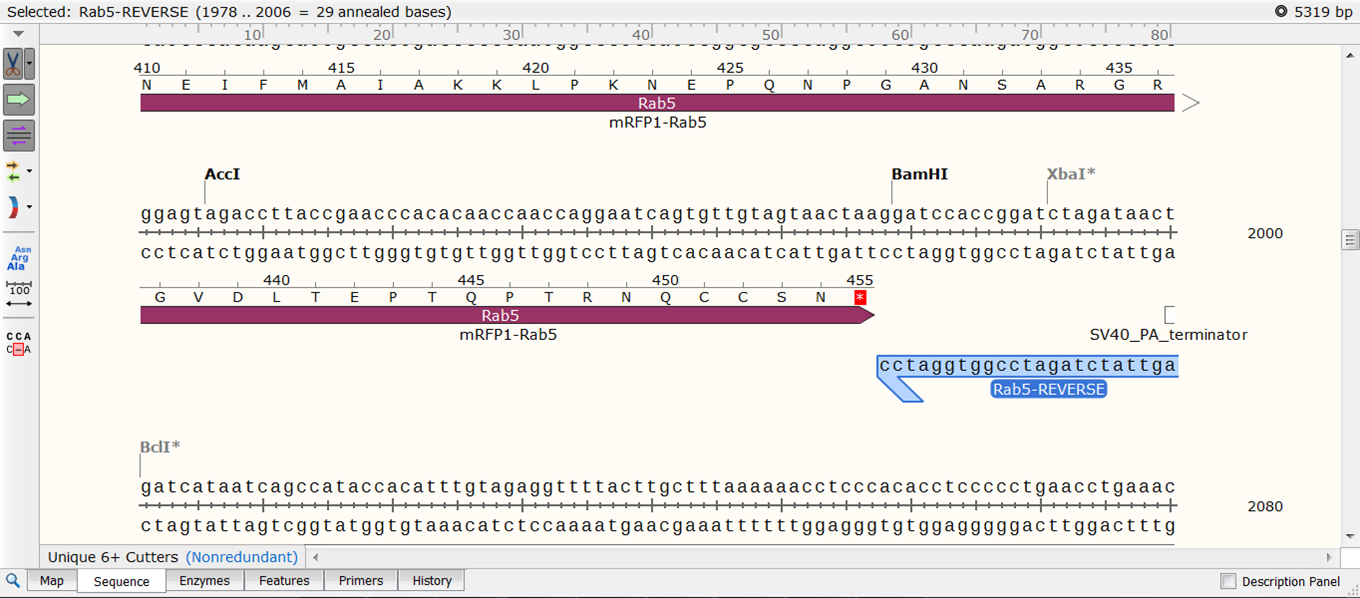
To transform the selection into a primer expand Primers in the top menu and select Add primer. Select to make a primer for the Bottom strand: The primer is displayed. Give it an informative name e.g. Rab5-REVERSE. If you are happy with the sequence of the primer click Add primer to template. |
If you want to design primers for other applications where efficiency and specificity of the primers can be taken into account, you shouldn't use SnapGene, use CLC Main Workbench or Primer-Blast instead.