Galaxy beginner's tutorial
Contents
- 1 Why Galaxy?
- 2 Before we begin
- 3 The analysis screen of Galaxy is divided in three sections
- 4 Getting data in
- 5 Datasets are collected in the history pane
- 6 Datasets are assigned to a specific type
- 7 Histories and their datasets are automatically saved
- 8 Security of your datasets and histories
- 9 Sharing histories
- 10 Running tools on your datasets
- 11 Galaxy 'DNA' workshop exercises
Why Galaxy?
Because of the tools. Because of the data storage. Because of the computing power. Because of the reproducibility of your research. Because you don't want to install all the tools yourself that are available on our Galaxy.
In following tutorial, we will guide you in exploring Galaxy for your bioinformatics needs. If the Galaxy of BITS does not suffice your needs, please contact us via bits@vib.be and we will get into touch whether we can meet them.
Before we begin
Before we begin, you have to log in on our Galaxy. If you don't have a login, please email bits@vib.be to obtain one.
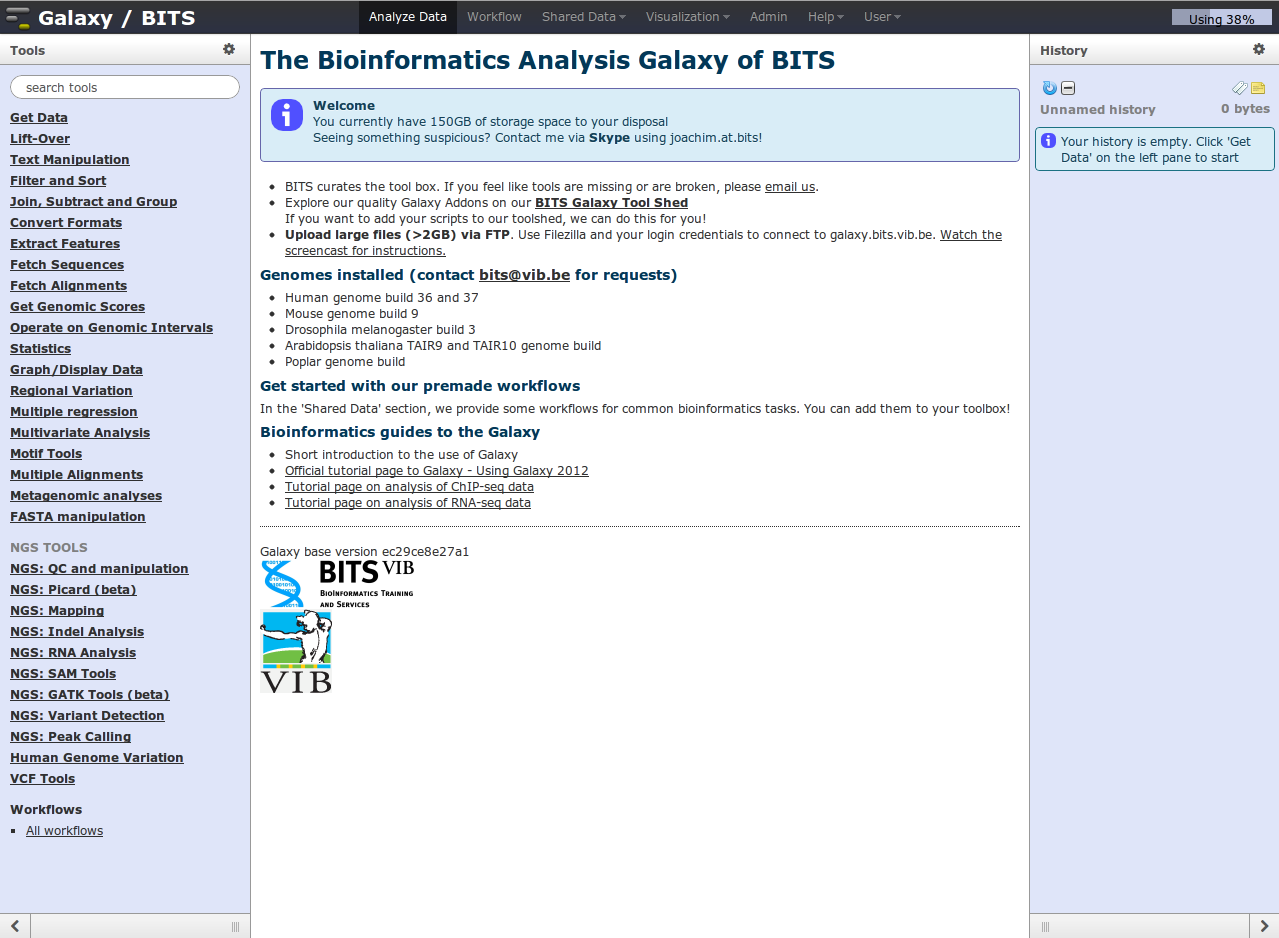
The analysis screen of Galaxy is divided in three sections
Tools at the left side, the middle pane in which the analysis results can be viewed, and the history on the right side showing the outputs of all previously run tools.
The black bar at the top provides an access point to more functionality, covered later on. In the top right corner you can see your storage usage as a bar.
Getting data in
Upload small (< 2GB) files from your computer
Do this via the tool 'Get data' and 'Upload file'.
Choose the file under 'File:' and click execute. Galaxy will try to automatically detect the type of file you have uploaded.
The dataset appears in the history at the right pane (as output of the 'Upload data' tool).
Upload big files (> 2GB) using FTP
To send big files (>2GB) you must use FTP instead of the browser. This is a two-steps process. We use the ftp client Filezilla to do this. You can install Filezilla on your computer via your software manager (or 'sudo apt-get install filezilla' on Ubuntu) (see www.http://filezilla-project.org/). Connect with Filezilla to galaxy.bits.vib.be with your Galaxy email as username and your password.
At the left pane of Filezilla, contents on your local computer is visible. At the right, the directory at Galaxy is visible. Navigate to the file on your computer you want to upload, double-click on it and the upload should start.
When the upload is finished, go in Galaxy to 'Get data' -> 'Upload file' to get the data finally into Galaxy. The upload file(s) are shown in the section 'Files uploaded via FTP' of this tool. Select the ones you want to import and click on execute.
The dataset appears in the history at the right pane (as output of the 'Upload data' tool).
Get data in from a webservice
You can directly fetch data from a webservice as UCSC or Biomart. These are listed under 'Get data'. The site will appear in the middle pane of Galaxy: set the filters right and choose the 'Send file to Galaxy' option. The exact approach differs between tools. After this, the dataset should be visible in your history.
Datasets are collected in the history pane
Give your history a good name
It is good practice from the moment a dataset it present, to give your history an appropriate name. Just click on the title, type the new name and hit enter.
The different colors of datasets represent different states
Datasets in your history are in one of the five states:
- Error creating thumbnail: Unable to save thumbnail to destinationUploading is busy
- Error creating thumbnail: Unable to save thumbnail to destinationA tool is queued: the resulting dataset does not exist yet
- Error creating thumbnail: Unable to save thumbnail to destinationA tool is running: the resulting data is being generated
- Error creating thumbnail: Unable to save thumbnail to destinationA tool has finished: the dataset is ready
- Error creating thumbnail: Unable to save thumbnail to destinationA tool has encountered an error: the dataset is not to trust
Clicking on the title of a dataset expose some details of the dataset
Clicking on the name of the dataset in the history unfolds a preview and some tools to manipulate the data.
- Error creating thumbnail: Unable to save thumbnail to destination- displays the content in the middle pane
- Error creating thumbnail: Unable to save thumbnail to destination- edit the metadata (attributes) of the dataset, e.g. the type of data. Important!
- Error creating thumbnail: Unable to save thumbnail to destination- deletes the dataset
- Error creating thumbnail: Unable to save thumbnail to destination- download the dataset to your computer
- Error creating thumbnail: Unable to save thumbnail to destination- view more info, e.g. which parameters where used for generating the data
- Error creating thumbnail: Unable to save thumbnail to destination- run this job again, displaying the tool's settings in the middle pane. Important!
The following are only applicable for data which can be viewed in genomic context.
- Error creating thumbnail: Unable to save thumbnail to destination- view in Trackster, the build-in genome browser
- Error creating thumbnail: Unable to save thumbnail to destination- tag the dataset, so you can easily retrieve it by searching
- Error creating thumbnail: Unable to save thumbnail to destination- add comments to the dataset, especially useful if sharing this dataset with colleagues.
Explore the dataset yourself at http://galaxy.bits.vib.be/u/joachim/h/basicprotocol2
Datasets are assigned to a specific type
Click onWithin Galaxy, two major dataset properties must be set. Most of the time, the tool that generates the dataset get this straight. With uploaded data, you might have to do this yourself: the datatype and the genome build it is related too (see screenshot below). In addition, it is good practice to change the name of the dataset here to something descriptive.
Data types are used to filter the input for tools: e.g. mappers only accept 'fastqsanger' dataset types.
Note above: some types (e.g. bed, interval,...) require that also the names of the columns are set. You can preview these names in the little preview window, and set them correct by clicking on
Histories and their datasets are automatically saved
You can start a new history anytime. You will not stop jobs currently running in any history! ClickChoosing Saved Histories shows all your histories, with the number of datasets, colour-coded according to the state the datasets are in. Here you can switch between different histories to view their datasets.
Security of your datasets and histories
All datasets generated by you belong to you by default. Galaxy allows setting permissions: on who can change access permissions, and on who can access datasets. Do this by clicking theThe upper box allow you to set who can change access permissions. The lower box is the allow access box. You can select here only so-called roles: roles are collections of users or groups of users. Only admins of Galaxy currently can create groups and roles, but you always have your own 'private' role, to which all your datasets belong by default. More info on security on Galaxy's Official wiki http://wiki.g2.bx.psu.edu/Learn/Security%20Features.
If sharing data with a user, permissions need to be adjusted, but luckily this can be done automatically.
Sharing histories
You can share histories with users. Click in the history menu on 'Publish or share'. You will see something similar as the screenshot below.
You can share in three ways:
- get a URL to your history, which you can email to other people (first button)
- get your history listed in the 'Published history' pages, listed under the top menu 'Shared Data'. (second button)
- share your history with (a) specific user(s) (third button)
Running tools on your datasets
Tools are organised in sections
You can browse the sections for the tool you need, or you can search the descriptions of the tools at the top.
Two main ways to run a tool
- click on the name of the tool in the left pane: the tool loads with default parameter settings.
- click on the 'Run this job again' button on a dataset Error creating thumbnail: Unable to save thumbnail to destination: the tool which created that dataset loads again with the same settings.
At the bottom you can find information about how to use the tools, and often also some examples.
Galaxy 'DNA' workshop exercises
Galaxy provides tools for all steps of 'next-gen' sequencing analysis. Analysis of sequencing data can be summarized as in the scheme depicted above. Below you can find tutorials to get you started with NGS analysis in Galaxy.
! Check your logins to access Galaxy for the Galaxy 'DNA' workshop
- Quality control of NGS data in Galaxy (Accompanying Galaxy history)
- Mapping of NGS data in Galaxy (Accompanying Galaxy history)
- Getting and manipulating genome tracks in Galaxy (Accompanying Galaxy history)