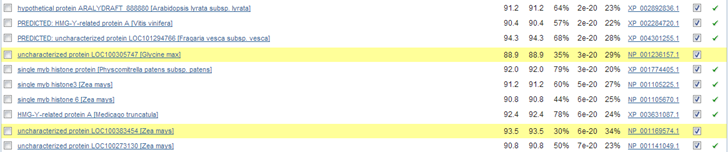Exercises on protein moltifs and domains
Go to parent Basic bioinformatics concepts, databases and tools#Exercises_during_the_training
Proteins exert their function through their specific 3D structure (tertiary and quaternary structure). Proteins that exert the same function often have similar (parts of) 3D structures since they're originally derived from the same ancestral protein. Similar parts of 3D structures correspond to a domain, which is a region in a protein that performs a specific function. Hence, when aligning proteins, stretches of similar AA residues (so called domains or motifs) can be detected. So, the first step to determine function of protein sequences is to look for known domains and motifs in the protein sequence that are related with a function.
Many webbased tools for this can be found on this page (and also on Expasy)
Contents
- 1 Searches for motifs or domains
- 1.1 Unknown motifs or domains
- 1.2 Known motifs and domains
- 1.2.1 Exercise 1: Ensembl
- 1.2.2 Exercise 2: NCBI's CCD
- 1.2.3 Exercise 3: PROSITE characterization of human TPA
- 1.2.4 Exercise 4: Retrieving all protein sequences containing a certain pattern
- 1.2.5 Exercise 5: search domains in human RIO2 via the InterPro database
- 1.2.6 Exercise 6: search domains in pig Q2L9D6 via Interpro
- 1.2.7 Exercise 7: Search domains in Bacillus subtilis O32142 with InterPro
Searches for motifs or domains
Unknown motifs or domains
Exercise 1: HMMER for building HMMs from protein alignments
HMMER is a very user-friendly tool for generating and using hidden markov models. A HMM can capture nearly all information (residues and gaps) of a multiple sequence alignment. When searching a database with a HMM, we can reach greater sensitivity than by searching the database with a single sequence as BLAST does. Use hmmsearch to search a protein sequence database using an alignment or a HMM
It only accepts a few MSA fortmats: often you need to convert your alignment to the right format. You can use a multiple sequence alignment converter.
Note that HMMER contains four programs:
- phmmer: to search a protein database with your protein sequence
- hmmscan: to search a HMM that matches your protein sequence
- hmmsearch: to search a protein database with your HMM
- jackhmmer: iterative procedure to search a protein sequenc e database with a protein sequence or HMM. After first iteration, the hits are added to the alignment, the HMM is refined and the search is repeated...
The hmmsearch option is what we will use. You should have on your computer 12 histone H1 protein sequences from plants (from the UniProt exercises. In the previous exercise we have aligned these sequences.
| Convert the alignment to a format that is accepted by HMMER. |
|---|
The resulting stockholm file is displayed in the "stockholm converted content" section. |
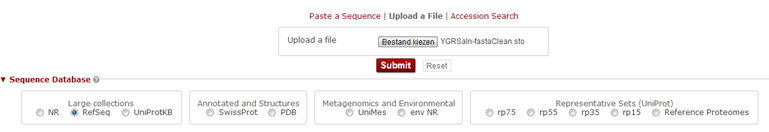
Now in hmmsearch select "Upload a File". Browse to the .sto file and click "Submit". Keep all default settings.
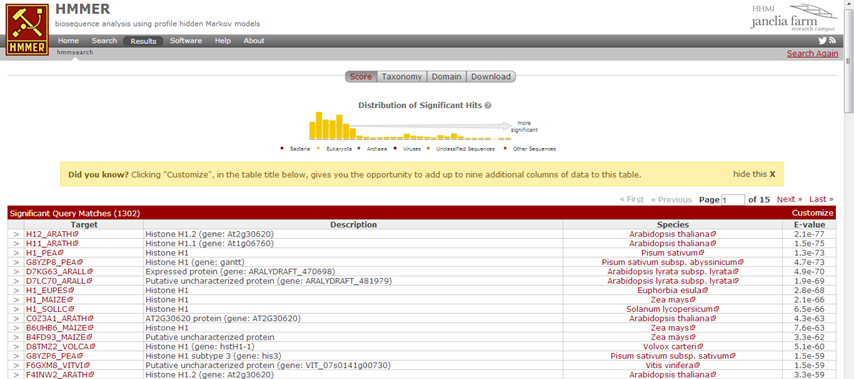
HMMER now creates a HMM of the alignment, and looks which protein sequences from UniProt (since we have selected to search against the UniProt database) match this HMM. Fairly quick, you get the results page showing the hits. Above you see a distribution of the E-values, and by hovering your mouse you can find more details about the contained hits. Such a profile is convenient to determine your threshold: if nice separate peaks exist, you can decide to set the threshold in the 'valley' between two peaks. Next in the results page a list of hits appears, with an E-value reported, similar as in BLAST.
| How many hits are reported by this search? |
|---|
| 1631 (as of June 2013) |
Hence, this search is an alternative to a simple BLAST search. We have created a HMM from sequences from Swissprot (hence high quality sequences) and next we have used this HMM to very sensitively search the UniprotKB for protein sequences that show similarity to the conserved parts of the proteins in our alignment.
Small note: the PSI-BLAST does a similar thing, but with a PSSM (position-specific scoring matrix) instead of a HMM.
Exercise 2: Searching for sequences with low similarity using PSI-BLAST
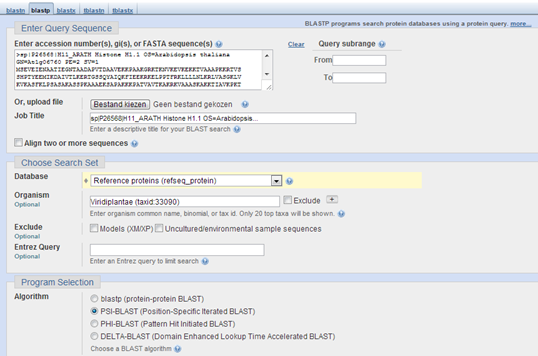
Use one of the histone H1 protein sequences from the exercise above about HMMER to search with PSI-BLAST to find similar sequences in the RefSeq protein database. I used H1.1 from Arabidopsis thaliana. Go to the BLAST page at NCBI and search with PSI-BLAST against Viridiplantae RefSeq protein sequences. Please note that PSI-BLAST searches can take a lot of time !
Training/tutorials on PSI-BLAST: http://sitemaker.umich.edu/microbial_genomics/files/psiblast3-3.doc
Known motifs and domains
Exercise 1: Ensembl
Information on the location of known motifs or domains in protein sequences can be found in many databases. One such example is Ensembl. Search the human F9 (Coagulation factor IX Precursor) gene and go to the transcript page of F9-001, its longest transcript (see exercises on Ensembl for details).
A nice link to try out is the "Display all genes with this domain" link, e.g. try it for the Peptidase_S1 domain.
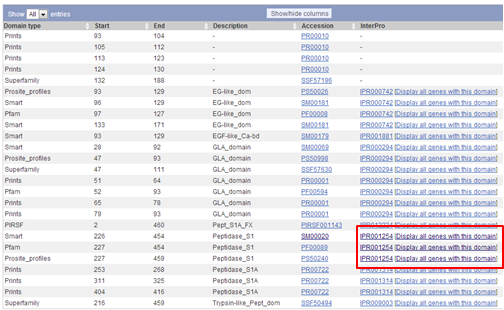
On the Transcript page you see now a graphical representation of all human chromosomes. On each chromosome, the positions of genes that encode a protein with a Peptidase_S1 domain is visualized by blue triangles. The location of F9 is shown as a red triangle.
Below this graphical representation, you see a list of all genes that encode a protein with a Peptidase_S1 domain. You can download this list by clicking on the Excel icon on the top of the list and selecting "Download whole table".
Exercise 2: NCBI's CCD
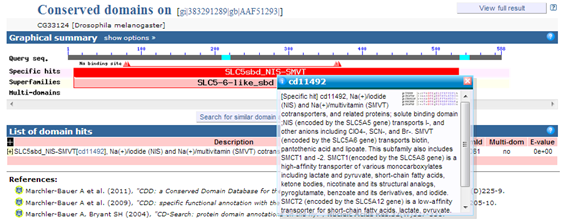
Search for protein AAF51293 on NCBI's Protein database.
Exercise 3: PROSITE characterization of human TPA
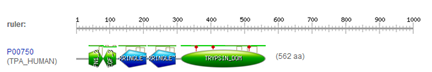
We have already analyzed the sequence of the human tissue type plasminogen activator (TPA) by downloading and aligning it in a dotplot. This protein has UniProt ID TPA_HUMAN and UniProt accession number P00750. This protein has well documented motifs, which we will look at using online tools
Of the many existing databases containing protein motifs and families PROSITE is the best curated and annotated. PROSITE allows you to:
- report the protein domains in your sequence
- report proteins containing a certain motif
We are going to analyze human TPA. In PROSITE you can provide an identifier instead of a sequence:
- Type 'tpa_human' in the bottom-left box under "Scan a sequence against PROSITE patterns and profiles"
- click "Scan"
Prosite will scan the sequence for all the patterns known in Prosite.
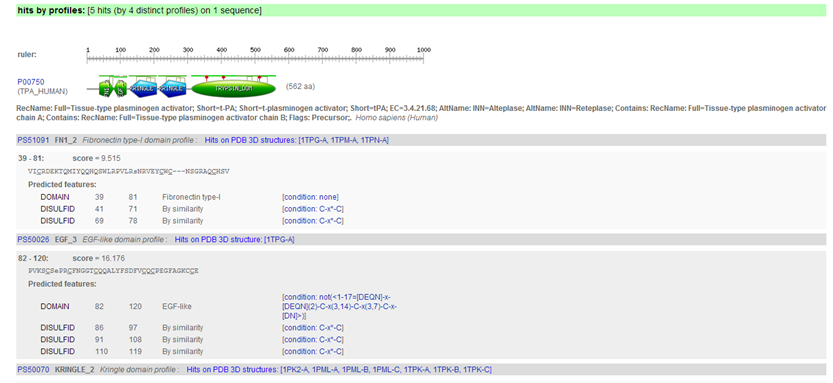
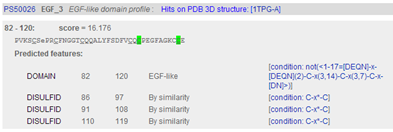
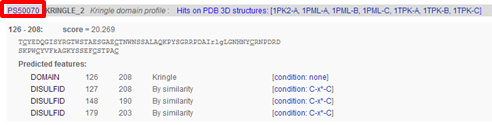
Results are returned in one long page. PROSITE contains motifs of two kinds: patterns and profiles.
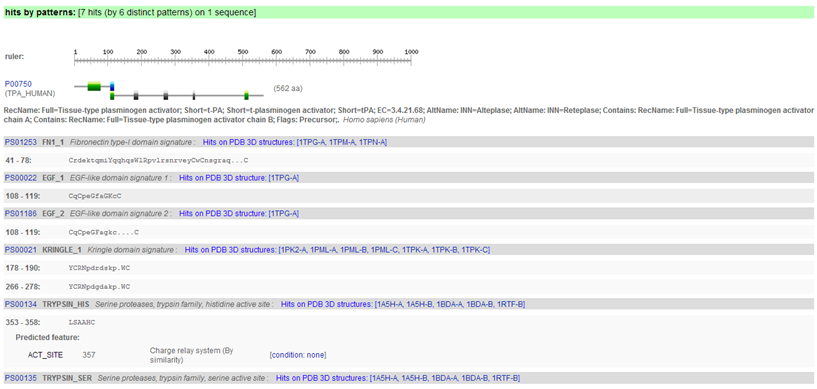
Hence, in the output page you will find first the "hits by profiles" (or "matrices") (usually longer motifs) and then the "hits by patterns" (usually shorter motifs).
The PROSITE search engine seems to have done a fine job in finding all the domains. A summary figure on top provides a nice overview.
The page is extensively postprocessed and provides a lot of extra information beyond the mere matches.
In the description of the individual motifs you can hover your mouse over certain components like "DISULFID" (C-C bridges) to highlight them in green in the domain sequence
In the complete sequence, the C's are highlighted in green and the domain in yellow.
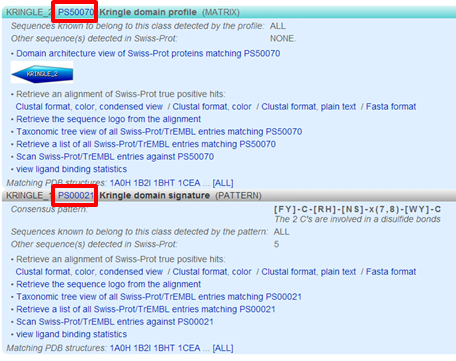
You can click on various links, e.g. click "PS50070"
to access the PROSITE documentation about kringles including a description of the domain and a list of proteins that contain the domain. From there you can click on "PS50070" (kringle domain) and "PS00021" (kringle motif) to access more detailed information, e.g. PSM of the domain, regular expression of the motif, list of UniProt sequences containing the domain/motif, list of sequences from PDB (for which the 3D structure was determined) containing the domain/motif.
Exercise 4: Retrieving all protein sequences containing a certain pattern
Prosite allows you to find all proteins that contain a certain pattern: the tool for this is called ScanProsite. The pattern must be written in a certain syntax, information on the syntax can be found here.
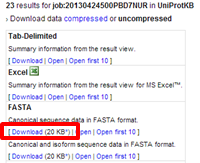
| Download all SwissProt sequences with motif YRGS at the C terminus in FASTA format as 'scanprosite_yrgs.fasta'. |
|---|
There are 23 SwissProt sequences containing this motif at their C-terminus. |
Please note that you can also search in one specific protein sequence for a motif. In that case you select Option 3.
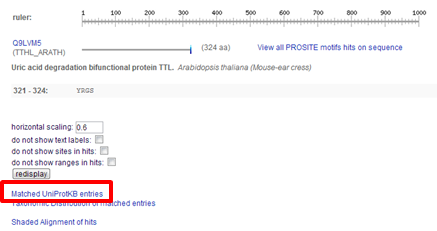
| Does the tpa_human protein contain the YRGS motif at its C-terminus. |
|---|
|
Let's get back to all protein sequences that do contain the C-terminal YRGS motif. You have downloaded them to a file 'scanprosite_yrgs.fasta'. Align the sequences.
| Which two are 'aberrant' ? |
|---|
| Use Muscle to create a multiple sequence alignment of the sequences. View the results in Jalview. In the alignment, you should see the YRGS motif at the C-terminus.
|
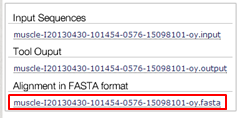
Go back to the FASTA file, remove these two sequences (this is easily done in WordPad) and align again.
Search with the above created alignment for similar sequences in the RefSeq database using HMMER.
| How many significant hits do you find ? |
|---|
| Download the alignment in FASTA format.
|
Exercise 5: search domains in human RIO2 via the InterPro database
InterPro is a composite database combining the information of all existing databases of protein families, motifs and domains. Some of these databases, like PROSITE, PRINTS and Pfam-A, contain motif and domain information. Other databases like ProDom and Pfam-B, contain information on protein families obtained via other methods (e.g. sequence clustering). The InterPro search engine searches the different member databases using their respective "native" search engines and then merges the results. Sometimes, Interpro finds motifs with different names from different databases that describe the same pattern: Interpro assembles them using an IPR-code!
Interpro works in the same two modes as Prosite does: you can search for a domain and retrieve all sequences that contain that domain or you can scan a sequence for an interpro domains (InterproScan).
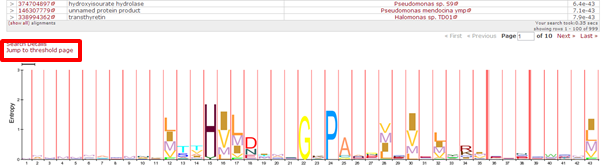
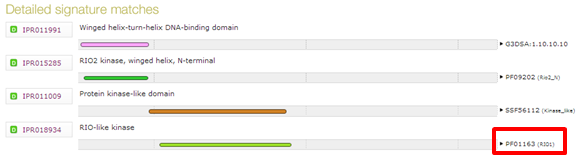
| Which domains do you find in human serine/threonine-protein kinase RIO2, with UniProt accession Q9BVS4 ? |
|---|
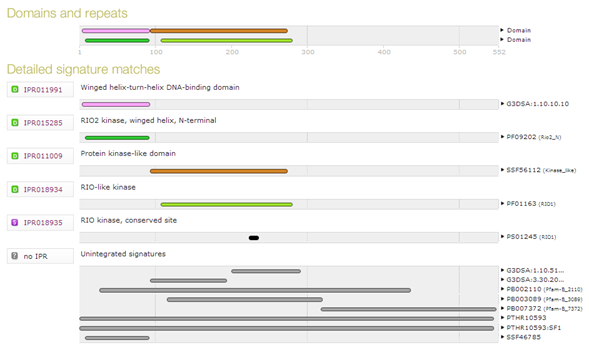 Which domain is correct ? They all are! |
| Which database does domain "IPR018934" come from ? |
|---|
When you go back to the InterPro page, please note the "Similar proteins" link in the left menu
When you click this link, you are redirected to a list of proteins with similar domain architecture. You can download the sequences of the proteins of this list in FASTA format by clicking the "Export FASTA" button on the top right of the page.
Exercise 6: search domains in pig Q2L9D6 via Interpro
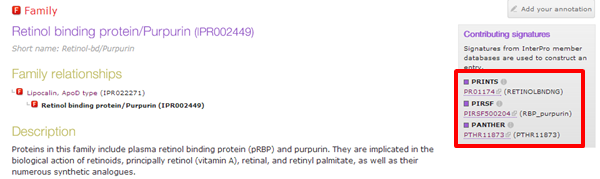
A very short protein from pigs has been found in blood (with UniProt accession Q2L9D6).
| What is the Interpro accession of the family this blood localized protein of pig belongs to ? |
|---|
| The family this protein belongs to is IPR002449. |
Exercise 7: Search domains in Bacillus subtilis O32142 with InterPro
Retrieve the sequence of Bacillus subtilis (O32142). Search for domains/motifs using InterProScan.
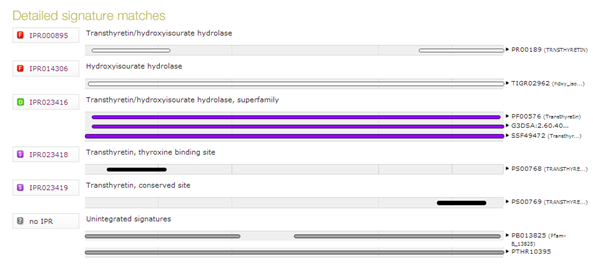
| How many domains/motifs does the sequence contain ? |
|---|
| You can use the Genbank accession to search Interpro. Type the accession number in the InterPro text search box and click "Search" |
| What is the relation between the two families that are returned by InterPro ? |
|---|
| Click on the InterPro ID of either of the two families to go to the InterPro record. There you can see that the second is a child of the first. |
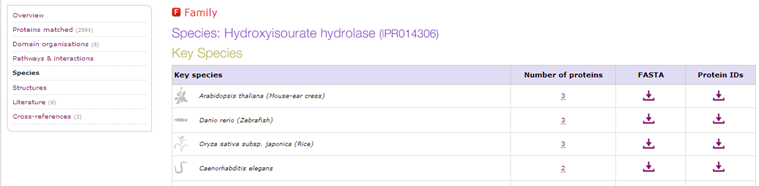
| Which species contain the most members of the child family ? |
|---|
| Go to the InterPro record of the child family. In the left menu click "Species". There you see that there are three members of the family in Arabidopsis, maize and zebrafish. |