Tophat
Tophat aligns RNA-seq data
Tophat is a mapper for aligning RNA-seq data to a reference genome (indexed with bowtie2), with the possibility to detect novel isoforms. It is optional to provide your own annotation files, or let Tophat detect novel isoforms by itself. For other aligner for RNA-seq, see http://wiki.bits.vib.be/index.php/RNAseq_toolbox#Aligning_of_RNA-seq_reads
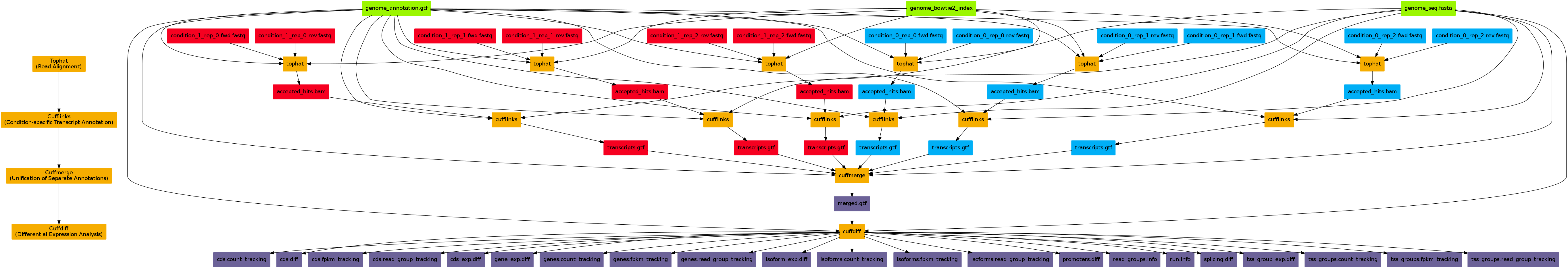
Tophat is the first step of the 'Tuxedo' suite of RNA-seq analysis tools for differential expression. The second step is cufflinks, which assembles an annotation file (.gtf) of the detected isoforms. The third step is cuffmerge, which merges all annotation of different mappings together. The fourth step, cuffdiff, takes this merged annotation file together with the mappings to estimate differential expression between the conditions.
See this figure for an overview of the Tuxedo suite:
Command examples
# a typical tophat command.
tophat -p 8 --no-coverage-search -o name_thout --transcriptome-index mm9.ensgene reads fq
In this example, 8 threads max are attributed to the job, coverage-search is avoided (speed), the file used to identify transcriptome is set to the ensemble gene model (other options are refGene of knownGene), finally, the read file is provided as input.
The result will be a folder named 'name_thout' and containing several files describing the result of mapping the reads to the ensgene exome.
-rwxrwx---. 1 root vboxsf 809M Oct 24 17:21 accepted_hits.bam -rwxrwx---. 1 root vboxsf 652K Oct 24 17:14 deletions.bed -rwxrwx---. 1 root vboxsf 329K Oct 24 17:14 insertions.bed -rwxrwx---. 1 root vboxsf 8.8M Oct 24 17:14 junctions.bed drwxrwx---. 1 root vboxsf 4.0K Oct 24 17:14 logs -rwxrwx---. 1 root vboxsf 70 Oct 24 09:48 prep_reads.info -rwxrwx---. 1 root vboxsf 17M Oct 24 17:14 unmapped.bam
Each group of reads is processed with tophat separately. This can take quite some time (up to ~10h on the BITS cluster for 15million reads).
The 'accepted_hits.bam' file is the input for the next processing step using cufflinks or other RNASeq quantification software (eg DESeq ...).