Literature Management
Contents
Summary
Executive summary
An average research article is full of literature references. Searching, storing, annotating, sharing and citing of scientific literature is an everyday task for every scientist. There exist an entire range of software tools, websites and methodologies to perform these tasks easier, faster and with more efficiency. However, it might be difficult to find the right tools with the features you as a scientist need to directly kick off. This training session helps you by showing some websites and software tools that you can directly start with to manage your literature collection.
Training schedule
The training is given by Maté Ongenaert. Maté is a PhD in Applied Biological Sciences, cell and genebiotechnology (2009). He currently is a postdoctoral researcher at the Center of Medical Genetics Ghent (CMGG).
Introduction
An introduction, showing the needs for efficient literature searching, annotation and storing with examples from both industrial and academic settings.
Searching literature
We will learn how to search literature in an efficient way, manually or automaticaly; once or at specified times.Therefore, we will use public websites such as PubMed/MedLine and Google Scholar.
- What:
- articles
- books
- patents
- official documents
- online resources
- Where:
- library
- Internet sources
- PubMed (myNCBI)
- Web of knowledge
- Google Scolar (iGoogle)
- EBI patents
- Espacenet
- Google patents
- Patentlens
- How
- manually
- automatically
- one time
- frequently
- focused area
- more general searches
Storing
One we have found the research articles we needed, we want to store their information (metadata): authors, title, dates,... There exist software tools that can generate such databases such as Reference Manager and Endnote. Of particular interest is a freely available literature managment program: Mendeley.
We will learn how to install and configure Mendeley and how to store literature references in it.
- Local
- Reference manager
- Endnote
- Zotero: in the Firefox browser, right where you need it!
- Mendeley: stand-alone literature managment program that deals with every aspect in storing: rename the pdf files (from article_1564686 to author_title or how you want it to be), organise your library,...
- Online
- Cite you like
Reading and sharing
You might want to read the fulltext version of the article. We'll show you how to get access to it, and save it along with the abstract and metadata. We will demonstrate the use of Mendeley to read articles, search in your collection and to manage and give structure your personal library.
Want to share the article with your colleages or collaborators? Some ways in exporting or sharing your very own library are demonstrated.
Get access to the full text article:
- link to pdf
- Google scolar
- contact the author
- SFX
Store PDF versions along with abstract and metadata:
- Mendeley
- ZotFile
Share your library
- Mendeley
Writing
It's time to start writing and citing! From within Word or OpenOffice/LibreOffice, access your library and generate in-text citations and bibliography in the style of the journal you are going to publish in. Article rejected? Change of citation style won't increase the pain...
- Try open access and your impact will improve !
Training (explanations and exercises)
Literature searching
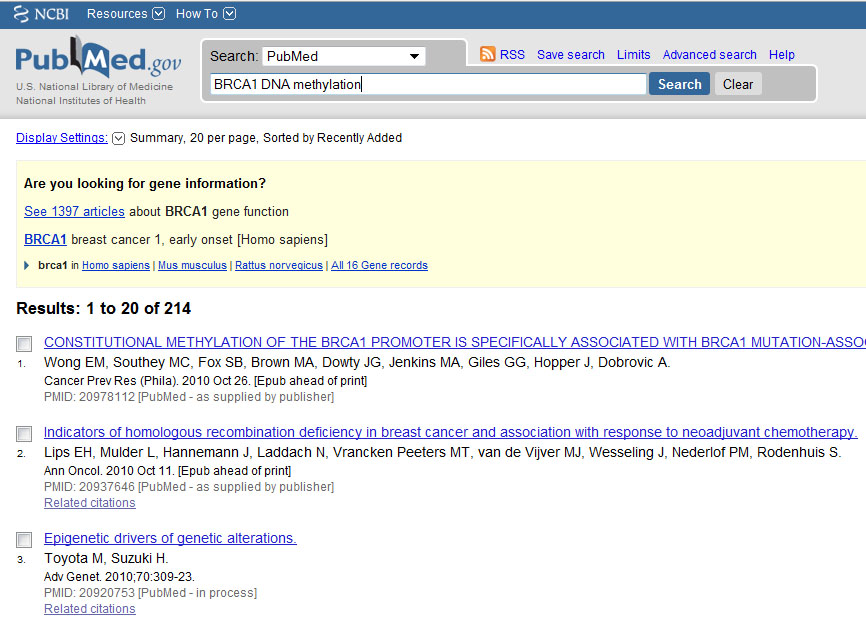
NCBI PubMed searching
Manual search:
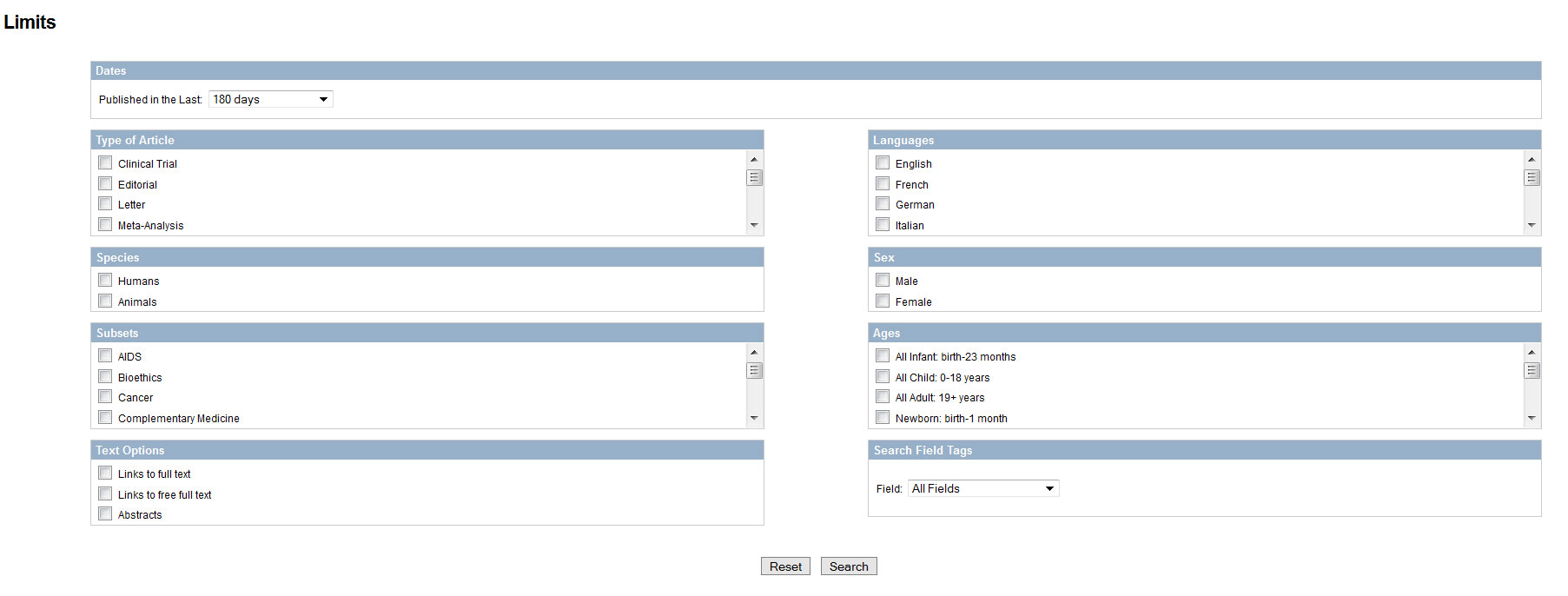
How does this looks like... Advanced search shows you a list of recent searches. Clikcing on the #... - details brings you to the details of your search.
Try to play with 'filters' and advanced serach options and see what happens with your search queries. Which fields (such as PDAT, MeSH Terms) are available within PubMed and how would you use them?
Great! Now we've got the results of our search and we have seen how our original query that is actually send to NCBI looks like.
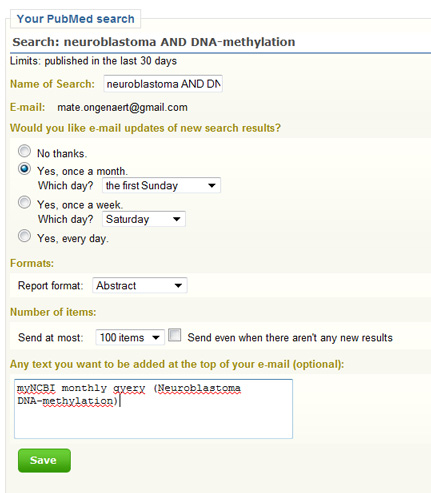
MyNCBI
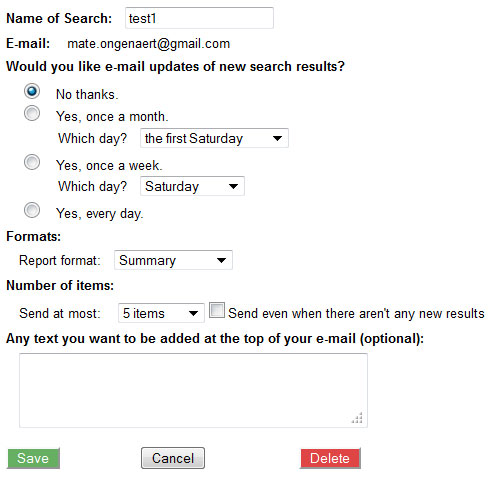
But what if I wanted to save this search, or execute this at regular times. For example: I want to keep track of all publications that deal with neuroblastoma and DNA-methylation. Therefor, I want to search PubMed every month and see the latest results. How can this be done? MyNCBI is created for saving and executing queries at regular times. How to proceed?
- go to myNCBI
- create an account or sign in trough a partner (Google etc.)
- save a search
- choose options such as execute every 30 days; get updates per email
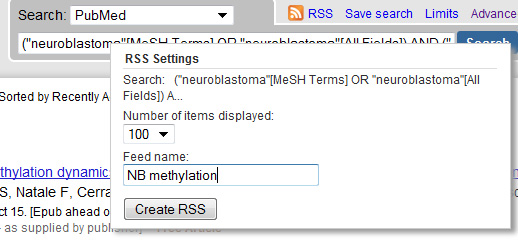
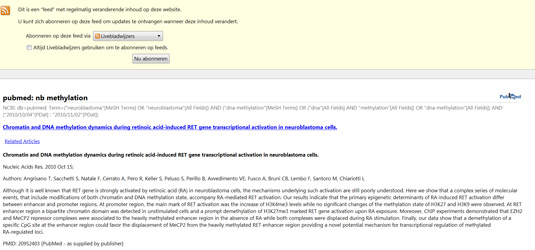
Other ways to represent the search? Want to share your carefully created search queries with your colleages, for example by showing the results on the intranet website? Maybe a RSS-feed is suitable! The RSS button shows you the options...
How does the result looks like in firefox?
Try creating your own searches at regular times and create an RSS feed to share with your colleages. Find out how RSS works and how the XML file behind it, is structured.
Also: there is a screencast about the above discussed features of PubMed: - PubMed screencast: [1]
Google Scholar searching
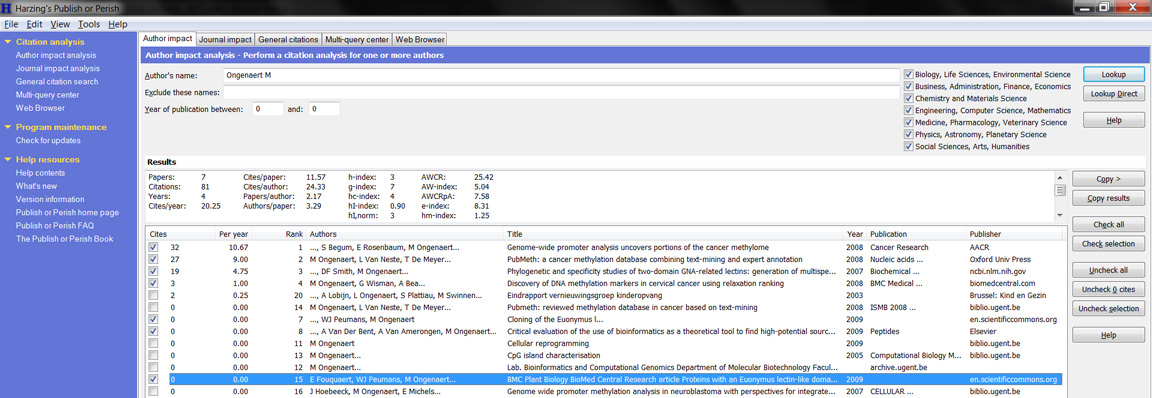
Try out Google Scholar and explore how you can create an email notice for new articles. What features are different from NCBI? What is better in Google compared to NCBI and what is rather worse? Useful feature is the view of the number of citations. Other applications use the information from Google Scholar, such as Publish or Perish, that shows statistics (number of cites, h-index,...) about an author or journal, based on a Scholar search.
Literature storing, reading and citing
Retrieve papers
It is important to share references, using unique identifiers. There are several of them, all with their own properties:
- PMID: unique ID in PubMed (NCBI)
- DOI: digital object identifier: unique identification for any file on the Internet, permanently available (permalink)
Retrieve your paper:
- PubMed
- Google Scholar
- Trough SFX (institution has to support this)
- Site of the publisher
- Ask the author
The importance of open access: makes sure that your article is freely available, not depending on a subscription of the possible readers. Will increase the impact of your research!
Some more advanced things
There exist a whole bunch of literature searching and mining tools. Most are based on:
- Query input and translation
- Query execution (to NCBI or others; local database)
- Result retrieval (for example NCBI E-Utils)
- Result interpretation (highlighting, sorting, counting, lookups,...)
- Visualisation population (e.g. co-occurence)
Some example tools are described in this training. I'll give a demonstration how to remotely call NCBI PubMed trough the E-Utils and how the results are returned. A similar mechanism is used in most of the presented tools. Afterwards, the data is analyzed. I'll also show how an example tool could be made, highlighting gene names and aliases and sorting according to relevance.
An overview of several existing online tools is given here: [2]
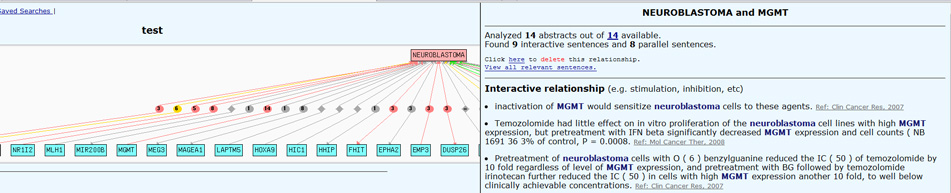
iHOP
Is rather a annotation-based tools; does not really allows mining but takes into account synonyms etc. and features highlighting and hover-over effects. [3]
GoPubMed
Retrieve articles with highlighting and hovering-over; see some visuals and statistics and see which GO-terms are present in the articles. [4]
PubGene
Create co-occurence networks between genes and biological terms [5]
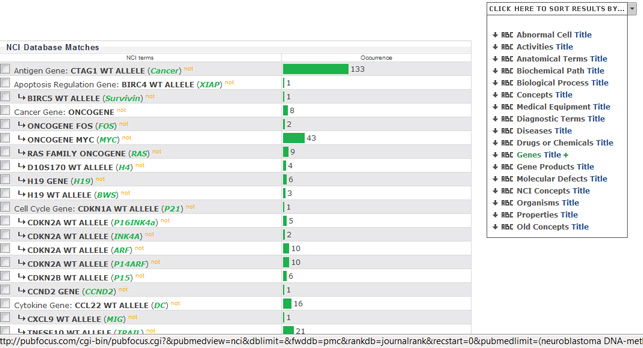
PubFocus
Try [6] to find genes names, associated with your query of interest. PubFocus translate your query to a PybMed search; executes it; retrieves and interprets the results. Visualisation is adapted to your choice.
Chilibot
Find associations between (and within) lists of keywords and genes, several search options are available. [7]
PolySearch
Find associations between diseases/genes/drugs/etc. [8]
Exercises
Exercises to complete - work in progress - should be ready before the training starts...
Exercise 1: PubMed searching
- Step-by-step exercises on the use of advanced searching options in PubMed
1. Make a query as you would usually do in PubMed
2. Modify your query, using operators (AND / OR / NOT) and several levels using brackets (e.g. neuroblastoma AND (methylation OR DNA-methylation)
3. Make use of the '*': using this symbol, PubMed will extend your queries (e.g. neuroblast* will be translated into neuroblastoma, neuroblasts,...)
4. On the top of the page, try setting some 'limits' (publication date, author, subject,...)
5. Try 'advanced', your search history will be shown, click on the #-symbol to see all options, select 'details' You will see the detail how your query is actually executed in full details (including MeSH terms) Fond out what MeSH terms exactly are and how you can use them.
6. Save your search (Save search on top of the Pubmed page) - you will be brought to the myNCBI website. Create a log-in or log in using your Google account. Save the search you just made and make an automated execution scheme (schedule your search)
- Step-by-step RSS exercise
1. On top of the pubmed search page, there is a 'RSS' option. Can you find out what RSS is and what the basic idea is behind RSS?
2. Create your own RSS feed, note its URL and check how it looks like in your browser or news reader
3. Try adding the RSS-feed you just created in Google Reader (google.com/reader)
Exercise 2: RSS feeds
1. Try to find the RSS-feeds of your favorite journal and add them as well. Find out how the RSS options of your browser / mail client work
2. (Advanced) Find how the XML-code behind an RSS feed looks like (hint: source code in your browser) and make your own RSS feed to share with colleagues,...
Exercise 3: Google scholar
1. Perform a search in Google scholar (scholar.google.com) and summarize what the differences are with searching PubMed
2. Try the 'advanced' search features in Google scholar
3. Set 'email alerts' for your search and compare with the email options from myNCBI and RSS feeds
4. What are the advantages and disadvantages of Scholar vs. Pubmed
5. (Advanced) 'Publish or Perish' is a tool that uses Google Scholar to extract biomedical information. Try out this tool (using an author or journal). Summarize how you think it might work to retrieve its information (draw as scheme how youy would create such a tool; start by exploring the Google Scholar options)
Exercise 4: Patents
1. By using EBI patents, Google patents, Esp@cenet or Patentlens, find if your gene of interest or your promoter/supervisor is mentioned in a patent
2. What is important in a patent search-tool and what are the differences between the above-mentioned search sites (hints: interface, full-text, advanced filtering, ability to search for sequences, search on company and person names)
Exercise 5: Mendeley
1. Register for mendeley (mendeley.com), download and install it
2. Once installed, follow the instructions in the instruction manual, that is in your library
3. What is crucial to have tested out are the following aspects: - starting from a PDF, get it in your library - web-importer: starting from PubMed/..., get the reference in your library - try the search-functions - read a PDF in Mendeley, highlight parts of it and add keywords or tags to the reference - set-up both a watched folder and a folder to manage your PDFs and think about you would use it - create a shared group, add some references and think about how you would use it - cite from within Word or openoffice
4. Import/export: starting from and EndNote library, how can you import this into Mendeley and how can you export to bibtex for a colleague you collaborate with
Exercise 6: NCBI E-Utils (advanced)
1. Find out how E-Utils work and what you can do with it
2. Which databases can you approach using NCBI E-Utils
3. Can you find a working E-Utils example in your favorite scripting/programming language? (Advanced)
4. Explain the difference between E-search and E-Fetch when using the PubMed database (Advanced)
5. Launch an E-utils query, using E-search and E-Fetch (Advanced)
Solutions
Solutions are given here (Slideshare): [9]