Introduction to relational databases and MySQL
From BITS wiki
This page accompanies the BITS training session "Introduction to relational databases and MySQL" (see BITS training page).
Exercises
| BioDB version 1. |
|---|
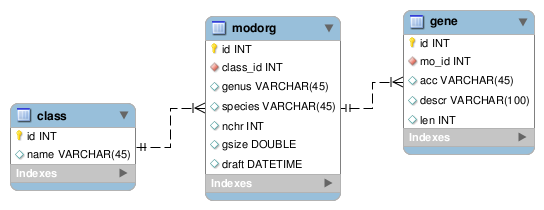
mysql> use biodb; Database changed mysql> select * from modorg; +----+------------+----------------+--------------+------+-------+------------+ | id | class | genus | species | nchr | gsize | draft | +----+------------+----------------+--------------+------+-------+------------+ | 1 | Bacteria | Escherichia | coli | 1 | 4.639 | 1997-09-05 | | 2 | Fungi | Saccharomyces | cerevisiae | 16 | 0.672 | 1996-10-25 | | 3 | Roundworms | Caenorhabditis | elegans | 6 | 100 | 1998-12-16 | | 4 | Insects | Drosophila | melanogester | 4 | 180 | 2000-03-25 | | 5 | Insects | Apis | melifera | 16 | 218 | 2003-12-19 | | 6 | Mammals | Mus | musculus | 21 | 2651 | 2010-09-23 | | 7 | Mammals | Rattus | norvegicus | 21 | 6597 | 2004-04-02 | | 8 | Mammals | Gorilla | gorilla | 24 | 3076 | 2008-11-03 | | 9 | Mammals | Homo | sapiens | 23 | 3038 | 2007-05-22 | | 10 | Aliens | Alienus | area51us | NULL | NULL | NULL | +----+------------+----------------+--------------+------+-------+------------+ 10 rows in set (0.00 sec) |
| BioDB version 2. |
|---|
mysql> use biodb; Database changed mysql> show tables; +-----------------+ | Tables_in_biodb | +-----------------+ | class | | gene | | modorg | +-----------------+ mysql> select * from class; +----+------------+ | id | name | +----+------------+ | 1 | Bacteria | | 2 | Fungi | | 3 | Roundworms | | 4 | Insects | | 5 | Mammals | | 6 | Aliens | +----+------------+ ysql> select * from modorg; +----+----------+----------------+--------------+------+-------+------------+ | id | class_id | genus | species | nchr | gsize | draft | +----+----------+----------------+--------------+------+-------+------------+ | 1 | 1 | Escherichia | coli | 1 | 4.639 | 1997-09-05 | | 2 | 2 | Saccharomyces | cerevisiae | 16 | 0.672 | 1996-10-25 | | 3 | 3 | Caenorhabditis | elegans | 6 | 100 | 1998-12-16 | | 4 | 4 | Drosophila | melanogester | 4 | 180 | 2000-03-25 | | 5 | 4 | Apis | melifera | 16 | 218 | 2003-12-19 | | 6 | 5 | Mus | musculus | 21 | 2651 | 2010-09-23 | | 7 | 5 | Rattus | norvegicus | 21 | 6597 | 2004-04-02 | | 8 | 5 | Gorilla | gorilla | 24 | 3076 | 2008-11-03 | | 9 | 5 | Homo | sapiens | 23 | 3038 | 2007-05-22 | | 10 | 6 | Alienus | area51us | NULL | NULL | NULL | +----+----------+----------------+--------------+------+-------+------------+ mysql> select id, mo_id, acc, len from gene; +----+-------+--------------+------+ | id | mo_id | acc | len | +----+-------+--------------+------+ | 1 | 9 | NM_000558 | 576 | | 2 | 9 | NM_000517 | 622 | | 3 | 9 | NM_000518 | 626 | | 4 | 9 | NM_000519 | 774 | | 5 | 6 | NM_008220 | 626 | | 6 | 6 | NM_008221 | 619 | | 7 | 6 | NM_008219 | 610 | | 8 | 7 | NM_033234 | 620 | | 9 | 7 | NM_001172845 | 589 | +----+-------+--------------+------+ mysql> select id, descr from gene; +----+--------------------------------------------------------------------+ | id | descr | +----+--------------------------------------------------------------------+ | 1 | Homo sapiens hemoglobin, alpha 1 (HBA1), mRNA | | 2 | Homo sapiens hemoglobin, alpha 2 (HBA2), mRNA | | 3 | Homo sapiens hemoglobin, beta (HBB), mRNA | | 4 | Homo sapiens hemoglobin, delta (HBD), mRNA | | 5 | Mus musculus hemoglobin, beta adult major chain (Hbb-b1), mRNA | | 6 | Mus musculus hemoglobin Y, beta-like embryonic chain (Hbb-y), mRNA | | 7 | Mus musculus hemoglobin Z, beta-like embryonic chain (Hbb-bh1) | | 8 | Rattus norvegicus hemoglobin, beta (Hbb), mRNA | | 9 | Rattus norvegicus hemoglobin, zeta (Hbz), mRNA | +----+--------------------------------------------------------------------+ |
| Solutions. |
|---|
Create database biodb
# mysql -p
mysql> create database biodb;
mysql> grant all on biodb.* to james@localhost;
$ mysql biodb < biodb1.sql
Navigation
$ mysql
mysql> show databases;
mysql> use biodb;
mysql> show tables;
mysql> show colums from modorg;
mysql> show create table modorg;
Sorting
mysql> select genus, species from modorg order by draft;
mysql> select genus, species from modorg order by nchr desc, genus, species;
Calculations
mysql> select concat(genus, " ", species), gsize / nchr, year(draft)
from modorg order by gsize / nchr desc;
Column aliases
mysql> select
concat(genus, " ", species) as name,
gsize / nchr as avgsize,
year(draft) as pubyear
from modorg
order by avgsize desc;
Conditions
mysql> select genus, species from modorg
where class = "mammals" and year(draft) >= 2005;
mysql> select genus, species from modorg
where gsize / nchr between 10 and 100;
mysql> select genus, species from modorg where genus >= 'a' and genus < 'f';
mysql> select genus, species from modorg where genus rlike '^[a-e]';
Limits
mysql> select distinct class from modorg order by class;
mysql> select genus, species from modorg order by gsize desc limit 3;
Count
mysql> select count(*) from modorg;
10
mysql> select count(nchr) from modorg;
9
mysql> select count(class) from modorg;
10
mysql> select count(distinct class) from modorg;
6
mysql> select count(*) from modorg where class = "mammals";
4
Groups
mysql> select class, count(*) from modorg group by class;
mysql> select class, min(gsize) as min, max(gsize) as max
from modorg
where gsize is not null
group by class
order by max desc;
Having
mysql> select class, avg(nchr) as avgchr
from modorg
group by class
having count(class) > 1
order by avgchr desc;
Joins
mysql> select name, genus, species, acc, len, descr
from gene, modorg, class
where gene.mo_id = modorg.id
and modorg.class_id = class.id;
Views
mysql> create view genevw as
select name, genus, species, acc, len, descr
from gene, modorg, class where gene.mo_id = modorg.id
and modorg.class_id = class.id;
mysql> select * from genevw where descr like "%hemoglobin%" order by len;
mysql> select min(len), max(len), avg(len), stddev(len)
from genevw
where descr like "%hemoglobin%"
|
Useful tips
| How to access a MySQL database from within a perl script? |
|---|
Make sure you have installed libdbi-perl and libdbd-mysql-perl in your linux box, then create a text file with following contents:
#!/usr/bin/perl -w
use strict;
use DBI;
my $dbh = DBI -> connect(
"DBI:mysql:host=localhost;database=biodb",
"joachim",
"joja2000") or die;
$dbh->disconnect();
|