Ex 7 Protein coding gene searching
Go to parent Introduction to Bioinformatics#Exercises_during_the_training
Contributed by Guy Bottu
GENSCAN
The EMBL entry with ID/AC X02419 contains the gene for the urokinase-type plasminogen activator. We will run some gene searching software against it and see if the software manages to correctly find the coding sequence. GENSCAN is one of the most popular tools for gene searching that does not rely on similarity with know genes. GENSCAN has been developed by Chris Burge and his group at the U. of Stanford. It is used by the genome annotation pipeline at the EMBL-EBI, while a slightly modified version (Gnomon) is used at the NCBI. There is a public server at the Massachusetts Institute of Technology.
GENSCAN models eukaryotic genes with a HMM (hidden markov model) and searches besides exons (or rather the coding parts of exons) also for the TATA-box, the CAP-site and the polyA-site. The public server offers models for human (probably usable for all vertebrates), Arabidopsis thaliana and maize.
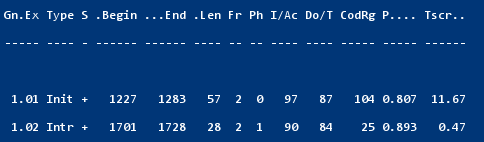
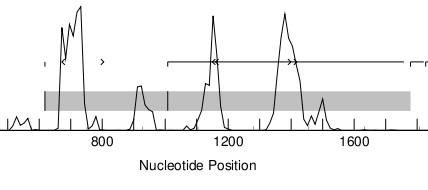
Go to http://genes.mit.edu/GENSCAN.html. Upload X02419.txt (the server only accepts "raw" sequence) and "Run GENSCAN". You can compare the result with the annotation in X02419.embl. You will see that GENSCAN correctly predicted the coding sequence and also found a polyA-site. Do note that GENSCAN does not even attempt to find non-coding exons.
GeneMark for eukaryotes
Another gene searching tool is GeneMark.HMM. It has been developed by Mark Borodovsky at the Georgia Institute of Technology and is commercialized by Gene Probe inc. GeneMark.HMM exists in a version for eukaryotes and a version for prokaryotes. Contrary to GENSCAN GeneMark.HMM eukaryotic does not try to find signals. GeneMark is interesting because it offers HMM models for a large variety of organisms.
Go to http://www.geneprobe.net and follow the links "About Us", "Georgia Institute of Technology", "GeneMark.hmm", "GeneMark.hmm for eukaryotes".
Upload X02419.fasta, set "Species" to "H.sapiens" and "Start GeneMark.hmm".
You will note that GeneMark did miss the last exon. This shows that gene prediction software is still not perfectly reliable. Do note that there exists a Genemark.HMM eukaryotic version 3 that does work fine on this gene ; it is not available on the public server but you can install it locally after you have obtained a licence (free for academics).
GeneMark for prokaryotes
GeneMark is one of the few gene searching tools that pays attention to prokaryotes. GeneMark.HMM prokaryotic is accompanied by a huge number of models for
different eubacteria and archaebacteria. For some species it also models the ribosome binding site (RBS). In case the organism you are working on (or a closely related
one) is not in the list, GeneMark has so-called "heuristic" models, based on genetic code and %GC. We will take as example the EMBL entry with ID/AC V00307. It contains the ompA gene (coding for a porin) and an ORF that has only much later after the sequence had been entered in EMBL/GenBank been shown to code for a component of the S.O.S.
system.
Go back to the page where you can choose between different versions of GeneMark.HMM and go to "GeneMark.hmm for prokaryotes". Do note how many
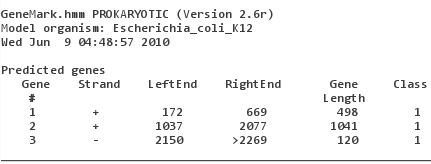
models for different species are available! Upload V00307.fasta, make sure that "Species" is set to "Escherichia_coli_K12" and "Start GeneMark.hmm".
You will note that GeneMark did find the two genes (and made a probably false-positive prediction of a reading frame running over the 3'-end). "Class 1" in the output refers to the fact that for most organisms GeneMark makes a difference between "typical genes" (class 1) and "atypical genes" (class 2).