Download and install the Kent tools, a collection of binaries and scripts
From BITS wiki
Go back to parent page Introduction to Linux for bioinformatics
Sometimes, the tool that you want is not available in a repository or the author does not provide installation files like a .deb file. No worries! You can also download the program from the internet. See below how!
The Kent NGS tools
The Kent tools are a collection of program files and scripts related to NGS data. It's freely available for you to use in your academic work. But there are no repo's or installation files available. We will install blat from Kent tools (blat is an extremely fast sequence aligner for very homologous sequences). It's available as an executable file online. You can download it, and simply run this file!
- Go to http://hgdownload.cse.ucsc.edu/admin/exe/linux.x86_64/
- Search for blat, and click on it.
- This opens the page with a download to the binary blat.
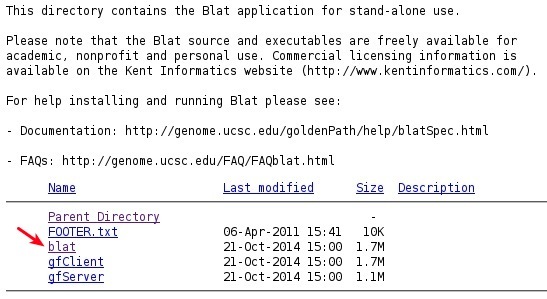
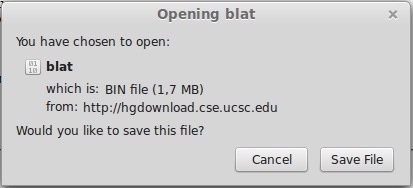
- Download the binary file.
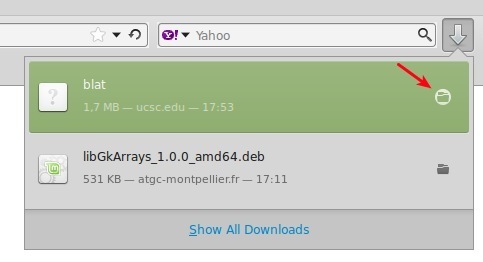
- Open the folder where the binary is saved.
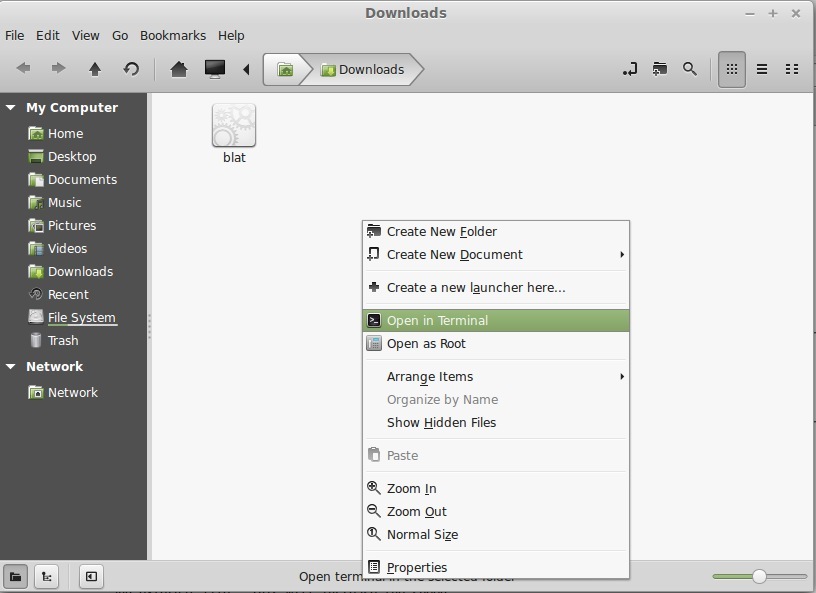
- Now, blat works via the terminal. We will cover the terminal in depth soon. For now, follow along: open the terminal by right-clicking in the folder
- Run the binary file 'blat' by typing in the terminal ./blat.
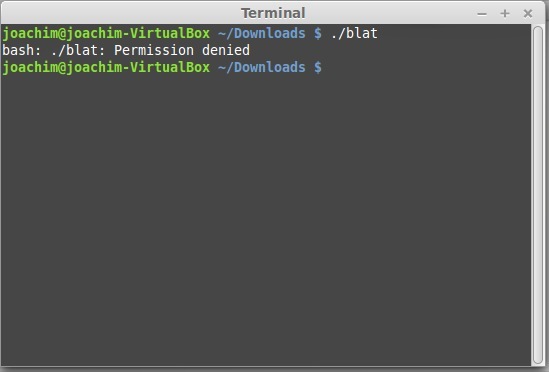
- The error you get, is because you are not allowed to run this! (this 'permission' we will see also later).
- To be able to run it, type in the terminal chmod +x blat.
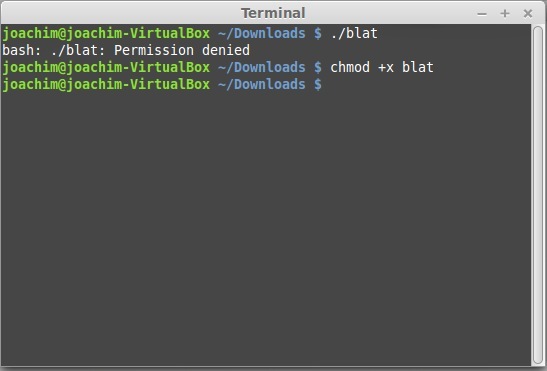
- Try to run blat again by typing ./blat.
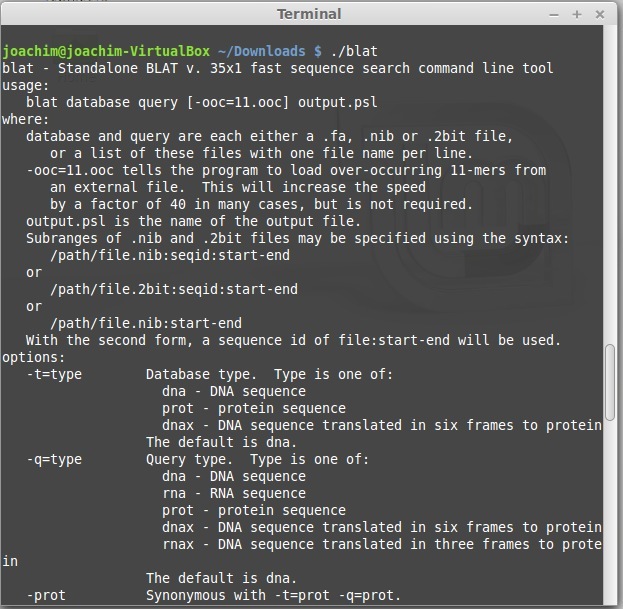
- Victory!
Tip! Make a separate 'bin' folder to store your binary files like 'blat'.
Go back to parent page Introduction to Linux for bioinformatics