... compare two long DNA strands for homologous regions
Finding the right tools to compare two long DNA stretches, is sometimes cumbersome. Here we show you how to use BLAST2SEQ and the Artemis comparison tool (ACT) to achieve this.
For example, we want to compare chromosome 1 and chromosome 8 of Saccharomyces cerevisiae. There seems to be something interesting going on over there.
1. First, we need to retrieve accession numbers (or sequences) for the chromosomes. In my opinion, following is the easiest way: go to Taxonomy at NCBI --> search S. cerevisiae --> click on genomic sequences --> search chromosome 1 and 18 (resp. accession number NC_001133 and NC_001140).
2. Then go to BLAST2SEQ, a modified blastn for blasting 2 streches of DNA. (you can access this page from the BLAST page on NCBI - at the bottom you will find blast2seq). Enter the accession numbers, each in one field. Depending on the origin of the sequences, select megablast or the discontiguous version.
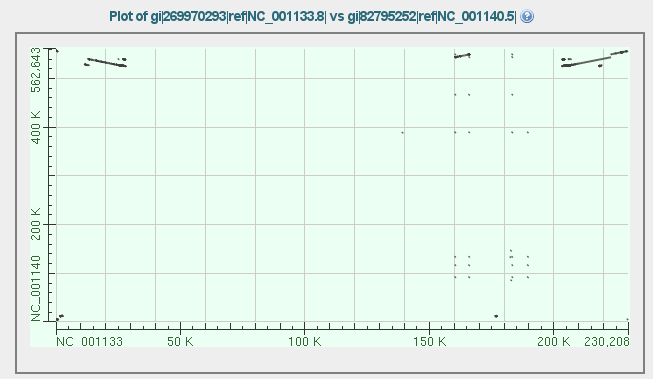
3. The two sequences are blasted against each other, after which a nice dotplot revealing you the positions of insertions (one reversed, three in 'same' direction). This is already a nice start to compare the stretches.
4. Download the text output from this BLAST2SEQ search as Alignment (HitTable(text)) on your computer.
5. MSPCrunch is a program to 'parse' BLAST output. You can accessible from Mobyle, a webportal with a lot of useful sequence manipulation and analysis tools (it contains also several dotplot programs. Search for MSPcrunch and open it. Enter the contents of the downloaded file, select 'Force blastn' and the '-d' option. Hit run and save the 'blast_output_data' as a text file.
![]()
![]()

6. Go to the ACT website, to the Downloads tab, and click on the Java webstart button. (JAVA needs to be installed. If needed, accept any certificates and allow running of the program)
![]()
7. Go to File -> Open... and enter both sequences (chr 1 and chr 8 in GenBank format), and as comparison file the MSPcrunch output.
8. Et voila, start to analyze your DNA sequences!
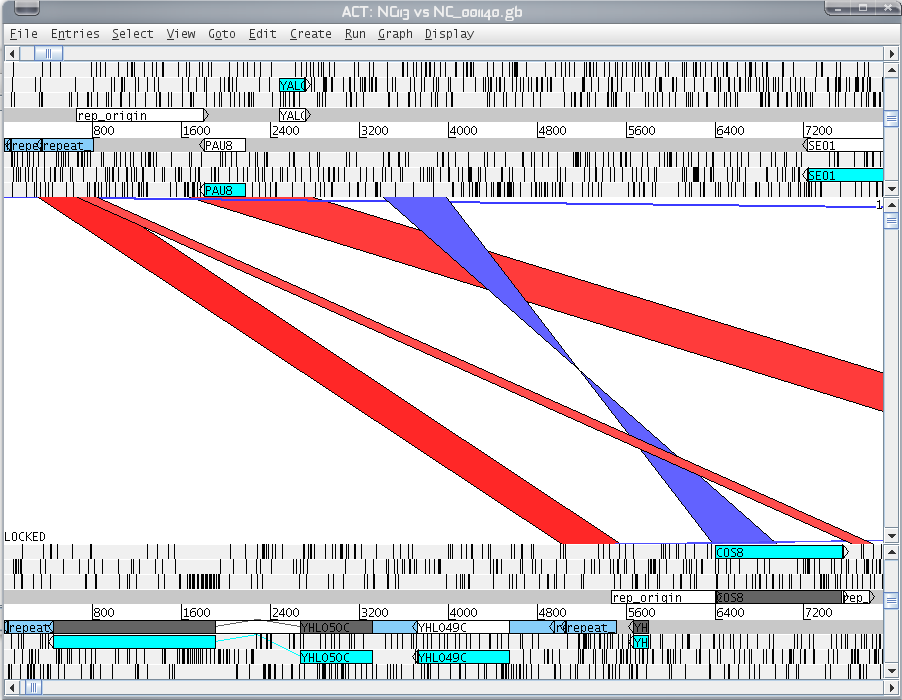
9. Now you know the positions of homologous regions: you could try to align them more specifically (clustalW, muscle).