Protein motifs and domains
Go to parent Basic bioinformatics concepts, databases and tools#Exercises_during_the_training
Proteins exert their function through their specific 3D structure. Proteins with the same function have similar structures because they are derived from the same ancestral protein. Conserved parts of 3D structures are called domains, regions in a protein that perform a specific function. So, the first step to determine the function of proteins is to look for known domains in the protein sequence that are related with a function.
Contents
- 1 Unknown motifs or domains
- 2 Known motifs and domains
- 2.1 *Ensembl domain information
- 2.2 *PROSITE characterization of human TPA
- 2.3 PROSITE: Retrieving all protein sequences containing a certain pattern
- 2.4 PROSITE: Checking if a protein sequence contains a certain pattern
- 2.5 *SMART: visualizing domain architecture in TRPA1_MOUSE
- 2.6 Interpro
- 2.7 Pfam
Unknown motifs or domains
HMMER: Searching homologs based on HMMs
HMMER is a set of tools that allows to generate hidden markov models for representing MSAs and use them for similarity searches. A HMM can capture all information (residues and gaps) of a MSA. When searching a database with a HMM, you can reach greater sensitivity than by searching the database with a single sequence like BLAST does.
The website contains four tools:
- hmmscan to search a sequence against a profile HMM database
- phmmer to search a sequence against a sequence datatbase. (BLAST-like)
- hmmsearch to search a sequence database for matches to a HMM.
- jackhmmer to iteratively search a sequence against a sequence database.(PSI-BLAST like)
*hmmscan: finding motifs and domains in a sequence
| Search human F9 (the golden transcript) against the Pfam database ? |
|---|
The output format is similar to that of BLAST. |
hmmsearch: finding sequences that match a HMM
HMMER is very picky about its input, only alignments in Stockholm format are accepted. Therefore, we will build the HMM in Ugene (which generates a HMM in a format that HMMER accepts) and then use hmmsearch to search a protein sequence database for sequences that are similar to the HMM.
| How to create a HMM in Ugene ? |
|---|
In the top menu:
 This opens the HMM3 Build window:
 Define an output file and click Build |
This will generate an output file containing the resulting HMM in the specified location. You can open these files in a text editor to take a look. The HMM is in a format that is accepted by hmmsearch.
| How to upload the HMM to hmmsearch ? |
|---|
|
Provide the input: the HMM created by Ugene. Click Upload a File, browse to the file of the HMM and open it. |
| How to do a search for homologs ? |
|---|
|
You have to define the database you want to search in.
HMMER uses two scores to measure similarity between a sequence and a HMM:
These scores have the same interpretation as in BLAST.
You can set a threshold for sequences and for hits. This is related to the fact that a HMM can match a sequence in multiple places. Each individual match gets a hit score and the sum of the hit scores of all matches on the same sequence is the sequence score. |
Fairly quick, you get a table of hits, with a link to the record of each hit in the database you searched in and an E-value, similar as in BLAST.

When you find potential novel homologs you can add them to the alignment to check if they fit the MSA. Often you will find annotation errors. This happens a lot: assembly and ab-initio gene prediction of genomes is done by an automated pipeline so many of the predictions are wrong.
To find homologs the HMM search is a much better tool than a simple BLAST search. You create a HMM from sequences from Swissprot (hence high quality sequences) and use this HMM to very sensitively search the Uniprot database for protein sequences that are similar to the conserved parts of the proteins in the alignment.
![]() In theory, PSI-BLAST does a similar analysis, but with a PSSM (position-specific scoring matrix) instead of a HMM
In theory, PSI-BLAST does a similar analysis, but with a PSSM (position-specific scoring matrix) instead of a HMM
PSI-BLAST: Searching homologs based on PSMs
Go to the BLAST page at NCBI and search with PSI-BLAST. Please note that PSI-BLAST searches can take a lot of time !
Position-Specific Iterative (PSI)-BLAST is a similarity search method that uses an alignment generated by a run of blastp to search a protein sequence database. The first iteration of PSI-BLAST is identical to a run of blastp.
| How to do the first round of PSI-BLAST ? |
|---|
Fill in the BLAST form:
|
When the results page appears, it looks similar to a regular BLAST search, apart from a small box above the results list.
This box is the power of PSI-BLAST: it allows to perform another BLAST search using a position score matrix constructed from the multiple alignment of selected hits of the blastp run. The matrix is used in place of the original substitution matrix for a second search of the database to detect sequences that match the conserved pattern specified by the PSM.
| How to do a second round of PSI-BLAST ? |
|---|
| Deselect hits you do not want to include in the alignment and click GO in the box above the results list. |
Iteration 2 will yield hits in the description table which are coloured in yellow meaning those hits were not found with the first search. You can continue to iterate this way, until no hits are labeled as new.

In this way PSI-BLAST can find more distantly related proteins.
Training/tutorials on PSI-BLAST: http://sitemaker.umich.edu/microbial_genomics/files/psiblast3-3.doc
Known motifs and domains
*Ensembl domain information
Information on the location of known motifs or domains in protein sequences can be found in many databases. One such example is Ensembl. Search the human F9 (Coagulation factor IX Precursor) gene and go to the transcript page of F9-001, its longest transcript (see exercises on Ensembl for details).
| In which part (N-terminal or C-terminal half) of the protein, encoded by F9-001, does the peptidase activity reside ? |
|---|
| On the transcript page, go to the left menu and click Domains & features.
 When you scroll down you see that the domains responsible for the peptidase activity are located in the C-terminal part of the protein. The peptidase activity is responsible for the cleavage of factor X to its active form factor Xa. |
A nice link to try out is the Display all genes with this domain link, e.g. try it for the Peptidase_S1 domain.
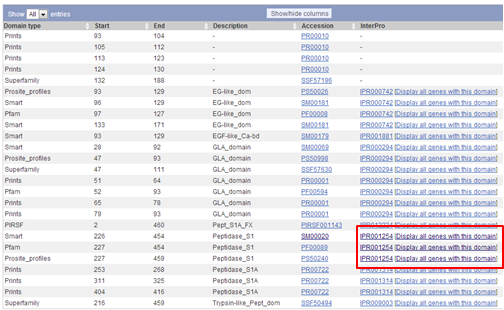
On the Transcript page you see now a graphical representation of all human chromosomes. On each chromosome, the positions of genes that encode a protein with a Peptidase_S1 domain is visualized by blue triangles. The location of F9 is shown as a red triangle.
Below this graphical representation, you see a list of all genes that encode a protein with a Peptidase_S1 domain. You can download this list by clicking on the Excel icon on the top of the list and selecting Download whole table.
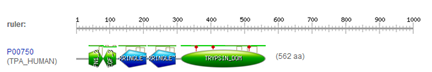
*PROSITE characterization of human TPA
We have already analyzed the sequence of the human tissue type plasminogen activator (TPA) by downloading it and aligning it in a dotplot. This protein has UniProt ID TPA_HUMAN and UniProt accession number P00750. This protein has well documented motifs, which we will look at using online tools.
There are many protein domain databases available but PROSITE is the best curated and annotated.
On the ScanPROSITE page you can perform various analyses using the information of the PROSITE database:
- Option 1: Search motifs from the PROSITE database in a query protein sequence: the input is a protein sequence, the output is a list of motifs.
- Option 2: Search sequences from a sequence database containing a query motif: the input is a motif, the output is a list of proteins containing the motif.
- Option 3: Search your set of sequences the ones that contain a query motif: the input is a motif and a list of sequences, the output is a list of sequences that contain the motif.
| Get a list of motifs that are present in tpa_human ? |
|---|
|

|
| Parameters of ScanProsite |
|---|
|
Note that you can select to
|
Results are returned in one long page. PROSITE contains motifs of two kinds: patterns and profiles.
Hence, in the output page you will find first the hits by profiles (domains represented by matrices) and then the hits by patterns (usually shorter motifs represented by regular expressions).
The page is extensively postprocessed and provides a lot of extra information beyond the mere matches.
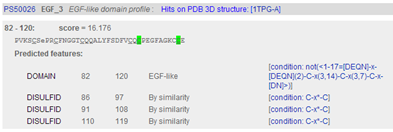
In the description of the individual motifs you can hover your mouse over certain components like DISULFID (C-C bridges) to highlight them in green in the domain sequence
In the complete sequence, the C's are highlighted in green and the domain in yellow.
| Which domain has the highest similarity score FN1 or EGF3 ? |
|---|
|
 |
These scores you are similarity scores, the sum of the individual scores of all positions in the alignment of motif and sequence.
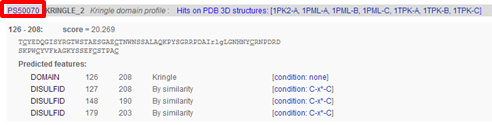
| Go to the Prosite record of the Kringle domain and check out the PSM of the domain |
|---|
|
Click the Prosite ID of the Kringle domain PS50070 to access the PROSITE documentation about kringles including a description of the domain and a list of proteins that contain the domain. From there you can click on PS50070 (kringle domain) and PS00021 (kringle motif) to access more detailed information, e.g. PSM of the domain, regular expression of the motif, list of UniProt sequences containing the domain/motif, list of sequences from PDB (for which the 3D structure was determined) containing the domain/motif.
 For instance when you click PS50070 you can view and download the PSM
 |
When you go back to the results page and you scroll down you see the list of patterns (motifs) found in TPA. The patterns don’t get similarity scores like the profiles (domains) but confidence levels. Since patterns are represented by regular expressions, these confidence levels can only have two values:
- 0: the sequence matches the regular expression
- 1: the sequence does not match the regular expression

PROSITE: Retrieving all protein sequences containing a certain pattern
Prosite allows you to find all proteins that contain a certain pattern: the tool for this is called ScanProsite. The pattern must be written in a certain syntax, information on the syntax can be found here.
| How to search protein sequences that contain a certain motif ? |
|---|
|
| How to download the hits found by ScanProsite ? |
|---|
|
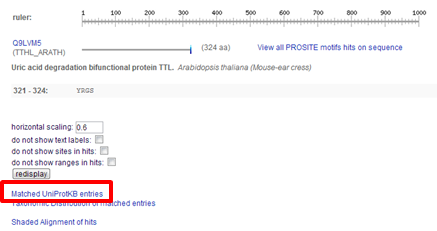
On the results page you can scroll to the bottom and click Matched UniProtKB entries In the screen that appears, all sequences but the isoforms are listed.
 |
PROSITE: Checking if a protein sequence contains a certain pattern
| How to search in a specific protein sequence for a specific motif ? |
|---|
|
In that case you select Option 3.
|
*SMART: visualizing domain architecture in TRPA1_MOUSE
The main focus of SMART is nice visualization of protein domains allowing to study the evolution of function within multidomain proteins. As an example we will visualize the domain architecture of TRPA1_MOUSE.
Go to the SMART website. You can use SMART in two different modes:
- Normal: the database contains Swiss-Prot, SP-TrEMBL and stable Ensembl proteomes
- Genomic: only the proteomes of completely sequenced genomes are used: Ensembl for metazoans and Swiss-Prot for the rest. See complete list of genomes in Genomic SMART.
We are going to use the Normal SMART. As you can see there are many ways to search SMART.
| Search the domain structure of TRPA1_MOUSE ? |
|---|
| Type TRPA1_MOUSE in the keywords box and hit search SMART to start the search. |
The search results in a nice visualization of the domain architecture of TRPA1_MOUSE and links to more in depth info which can help you determine the possible function of the protein.
Interpro
InterPro is a composite database combining the information of many databases of protein motifs and domains (see slides for overview). InterproScan, Interpro's search engine, searches all member databases using their respective "native" search engines and then merges the results. Sometimes, InterproScan finds motifs with different names from different databases that describe the same pattern.

Interpro works in the same two modes as Prosite does: you can search for a domain and retrieve all sequences that contain that domain or you can scan a sequence for an Interpro domain. In the latter case, you can search Interpro using the protein sequence (B) or via a text search (A e.g., with a word or short phrase, a UniProt or InterPro identifier).
| How to scan a protein for motifs and domains ? |
|---|
|
Once the analysis is complete, a results page will be returned showing the Interpro matches to your query sequence in a graphic. On the results page you find info on:
- Section A: shows the protein family to which Interpro predicts the sequence belongs.This is sometimes displayed as a hierarchy. Clicking the link will take you to the Interpro record for the family, where detailed information about its function may be found.
- Section B: a summary showing all domains and repeats that Interpro predicts the protein to contain. The protein is represented as a grey bar. Domains and repeats are indicated as coloured bars. Mousing over the bars reveals the type of domain or repeat, along with their position and a link to the relevant Interpro record.
- Section C: a detailed list of all families, domains, repeats and motifs the query protein contains. The information displayed in this section can be controlled using the interactive menu of Section D.
- Section E: shows the (GO) terms (see day 3) predicted for the protein. These terms are assigned based on the matches to the Interpro entries.

Each component is preceeded by an icon depicting its type (see Filter view on box in left menu for legend) and its Interpro accession number. At the left you see accession numbers/links to the member database the component comes from. Click the accession of the member database to go to the corresponding record in that database.
| How to download a list of proteins with similar domain architecture ? |
|---|
|
Click the Similar proteins link in the left menu: you are redirected to a list of proteins with similar domain architecture. You can download the sequences of the proteins of this list in FASTA format by clicking the Export FASTA button on the top right of the page. |
| How to go to the Interpro record of a motif or a domain ? |
|---|
|
Click the Interpro accession of the domain, the accession starts with IPR and is followed by 6 integers, on the InterPro results page. This will redirect you to the InterPro record of the domain. |
Interpro contains a lot additional info:
- The menu at the left contains links to more information about the domain: other proteins containing the domain, which species contain proteins that possess the domain, in which pathways are these proteins involved...
- The menu at the right contains links to the records of the domain in the member databases.
Often, people want to search Interpro with a batch of sequences e.g. to analyse whole proteomes. There are two ways in which it is possible to do this:
- Download and install the InterProScan analysis tool. The software comes with full installation instructions.
- Use EBI's web service. These allow up to 30 sequences to be analysed per request. Instructions on how to use the SOAP-based web service.
Pfam
Pfam provides high quality HMMs for all protein domains it contains. It builds it HMMs based on experimental evidence: proteins that are proven to have the same function. The HMMs can be downloaded and are the ideal starting point to search for more distant family members using HMMER. Remember HMMER is very picky about the format of the HMMs. Fortunately, Pfam offers HMMs in Stockholm format, the only format that is accepted by HMMER.
PfamScan allows to search a protein sequence for Pfam domains, very similar to InterproScan and ScanProsite.
| How to download a HMM in Stockholm format. |
|---|
|
Pfam itself has also done HMMER searches: as well as the seed alignment from which the family is built, they provide full alignments, generated by searching sequence databases using the family HMM, which is exactly what HMMER does.
You can visualize the Pfam alignment in Ugene via File -> Open (you have to tell him it's Stockholm format) or you can use it as input in HMMER.
Pfam also contains logos of the HMMs.
| How to look at the logo of a HMM ? |
|---|
|
Click the HMM logo link in the left menu. |
The combination of Pfam and HMMER is used to annotate proteomes of newly sequenced species. HMMs of Pfam used to find homologs in the proteome of the newly sequenced species using HMMER.