Predicting protein structure - homology modeling exercises
Exercises created by Joost Van Durme
Part of Protein Structure Analysis training
Contents
Introduction
The goal of homology modeling is to predict the 3D structure of a protein that comes close to what would be achieved experimentally with X-Ray experiments.
- Main principles of homology modeling
- We predict the structure of a protein sequence on the basis of the structure of another protein with a similar sequence (the template)
- If the sequences are similar, the structures will have a similar fold
- Structure is more conserved than sequence
The sequence
Last week my colleague sequenced a plant protein. He is not a bioinformatician and is actually scared of computers. Yet, he would like to know what the structure might look like to do some rounds of rational mutagenesis. I told him I would collect a group of bioinformaticians to solve this problem. Please don't disappoint him.
He came up with this sequence:
SVCCPSLVARTNYNVCRLPGTEAALCATFTGCIIIPGATCGGDYAN
Searching for the template structure
Well, actually, the first step is to check whether the PDB already contains the structure of this sequence. That would be easy so we don't have to model anything. We will use Blast again to search with the sequence.
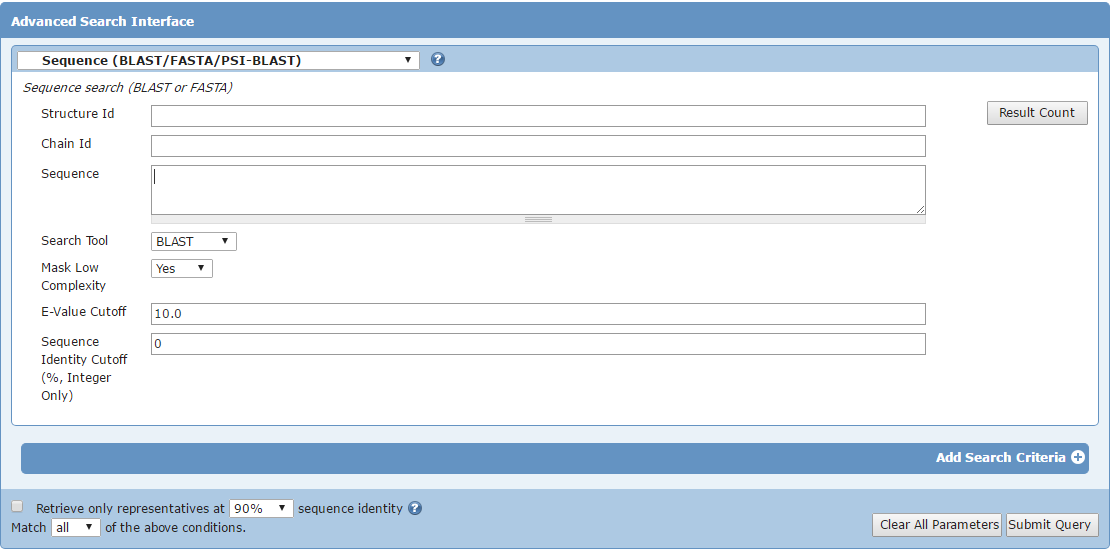
Go to the Advanced search button on the PDB home page at http://www.pdb.org and choose Blast/Fasta from the Query Type list
Copy and paste your sequence in the sequence field and submit query.
If the first hit is not a perfect 100% match, it means the structure of this particular sequence has not been solved and we have to build a homology model with a suitable template structure.
Click on Display Full Alignment per hit to see if the hit is a 100% match or not. By default, the most similar hits are at the top of the list (see the Blast E-value).
- A suitable template structure to make a high quality model should have following properties
- The highest possible sequence identity from all structures in the PDB when aligned to the target sequence
- A good resolution (and R-factor): if many identical template structures exist with the same sequence, filter by resolution
- Is solved by X-RAY, not NMR
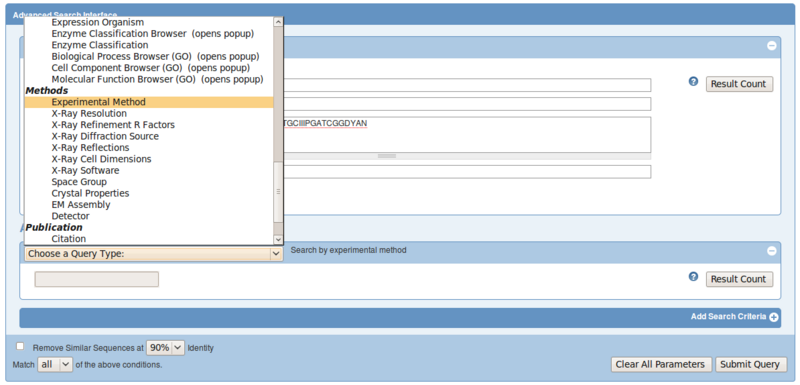
So let's search again with the sequence, but now filter out only structures solved by X-Ray:
- open the advanced search again, select Blast/Fasta and paste the sequence
A couple of structures show up now. Let's analyse the first 5 structures.
The results are sorted by BLAST E-Value, so closest homologs are shown first. And we want a close homolog as template structure.
We only want X-Ray structure, so skip NMR structures.
For each hit you can click on Display full alignment. Is there a difference in the number of identities, positives and gaps between the two remaining x-ray structures?
What is the PDB ID with the highest resolution, does not have insertions or deletions and should thus be the better template structure?
- Intermezzo
- You may also use other Blast servers such as:
- http://blast.ncbi.nlm.nih.gov/Blast.cgi
- Just make sure you are always Blasting against the PDB database.
- These servers might give you the results in a slightly different order, but as long as you follow the rules for choosing a suitable template, you should be safe.
Aligning target and template sequence and template selection
The alignment is the most crucial part of homology modeling. I will not explain what an alignment is and how you make it, this should be known. In an alignment we put homologous sequences on top of each other in a text file. The point is that amino acids that are on top of each other in the same column are assumed to have the equivalent position in the 3D structure. So if the template sequence has an Ala at position 3, where the target sequence has a Phe, homology modelling tools will use the backbone of the template structure and replace the sidechain at position 3 from Ala to Phe.
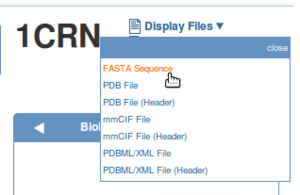
To extract the sequence from the template structure, display the FASTA sequence of the template structure and paste it into the text editor on the first line.
Open a text editor and copy and paste the template sequence on the first line and the target sequence on the second line like this:
TTCCPSIVARSNFNVCRLPGTSEALCATYTGCIIIPGATCPGDYAN SVCCPSLVARTNYNVCRLPGTEAALCATFTGCIIIPGATCGGDYAN
And save the text file.
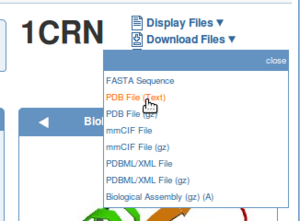
Also download the PDB file of the template structure from the PDB or directly in YASARA. In this example, we could choose 1CRN as template.
Homology modelling evolved over the years and many online tools for homology modelling are available. In my experience, homology modelling can be rather difficult and needs expert knowledge depending on the actual situation (sequence conservation, available templates, etc.). Below, you can find the procedure which has been used in FoldX version 3. I leave the description alive but we will not do this part of the exercise since in the newer vesion of FoldX this feature is no longer available.
Can you imagine what could be the reasons?
Building the homology model with FoldX version 3
The part of the exercise is only available for reference. We can no longer execute it.
We now have everything we need to build a model with FoldX version 3:
- Alignment of template and target sequence
- Template structure
Open the template structure in YASARA and choose a nice view. My favorite is F5 followed by F8.
It is very important to first energy minimize the side chains (Repair), so that FoldX makes an accurate model. Minimize the structure with:
Analyze > FoldX > Repair object
And choose the only object (object 1) in the left column.
Build the model by going to:
Analyze > FoldX > Build homology model
A series of menus are presented and this is what you should do:
- select the Repaired template Object: normally this is Object 2 from the left Sequence column
- select the alignment file you just made
- Move neigbours and Repair template object should be selected in the first options menu
- the numerical options should not be changed in the second options menu
Wait and you shall see your homology model of the target sequence as Object 3.
Now do a stability analysis (free energy calculation) on both the Repaired template structure and the model:
Analyze > FoldX > Stability of object
Select both the Repaired object and the model (normally Objects 2 and 3 in the left Sequence column).
Which one is more stable in terms of structure stability?
Building the homology model with Swiss Model
Our current request for homology is a rather safe one, so we can use an automatic server for homology modelling. There are many automatic tools available and many of them compete in regular competitions like lastly, the 12th Community Wide Experiment on the Critical Assessment of Techniques for Protein Structure Prediction (CASP12) - [1].
In our example, we take the Swiss Model server at [2]. SWISS-MODEL is a fully automated protein structure homology-modelling server, accessible via the ExPASy web server, or from the program DeepView (Swiss Pdb-Viewer). The purpose of this server is to make Protein Modelling accessible to all biochemists and molecular biologists worldwide.
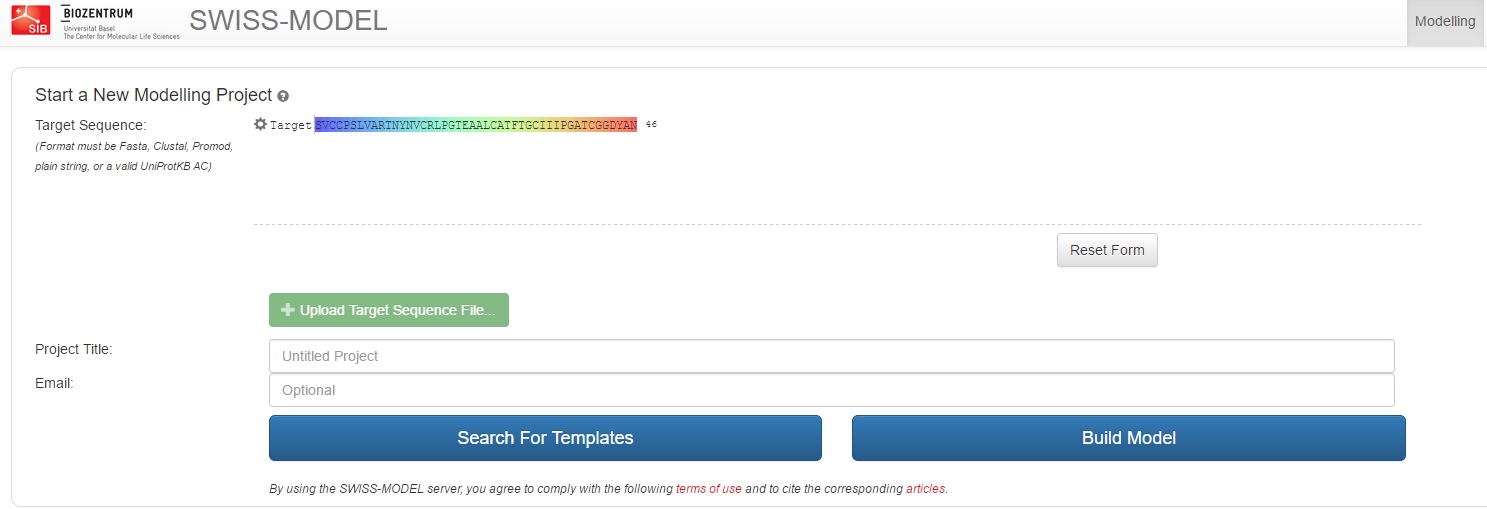
On the first page, after hitting Start Modelling, paste the sequence of our unknown protein in the field 'Target Sequence' and give the project a name. Click 'Search templates' to initiate the first step.
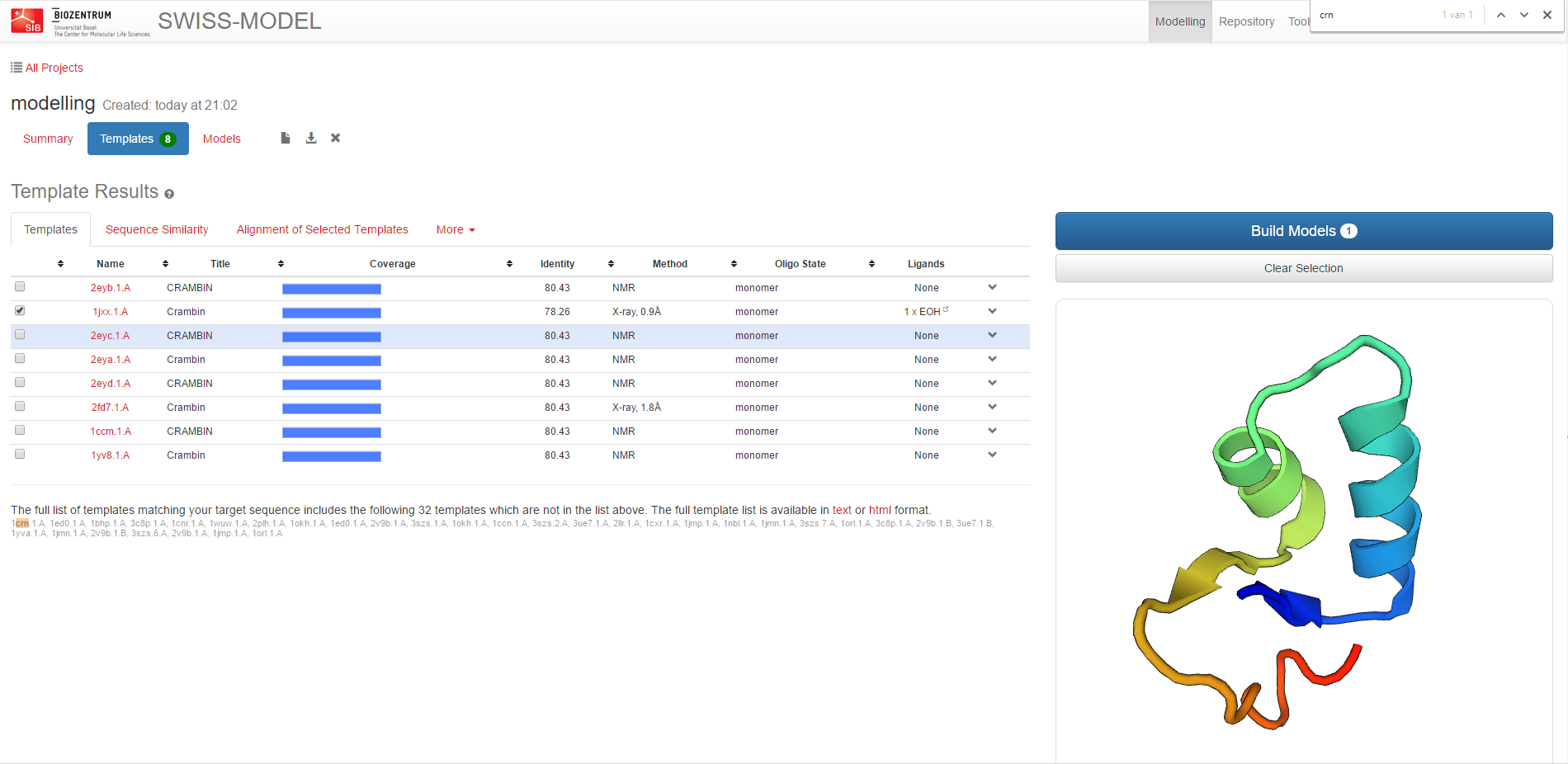
Thereafter, the server identifies structural template(s) and gives an overview list of hits which you can select the templates from.
After careful selection of the template, launch the Model building step.
Which
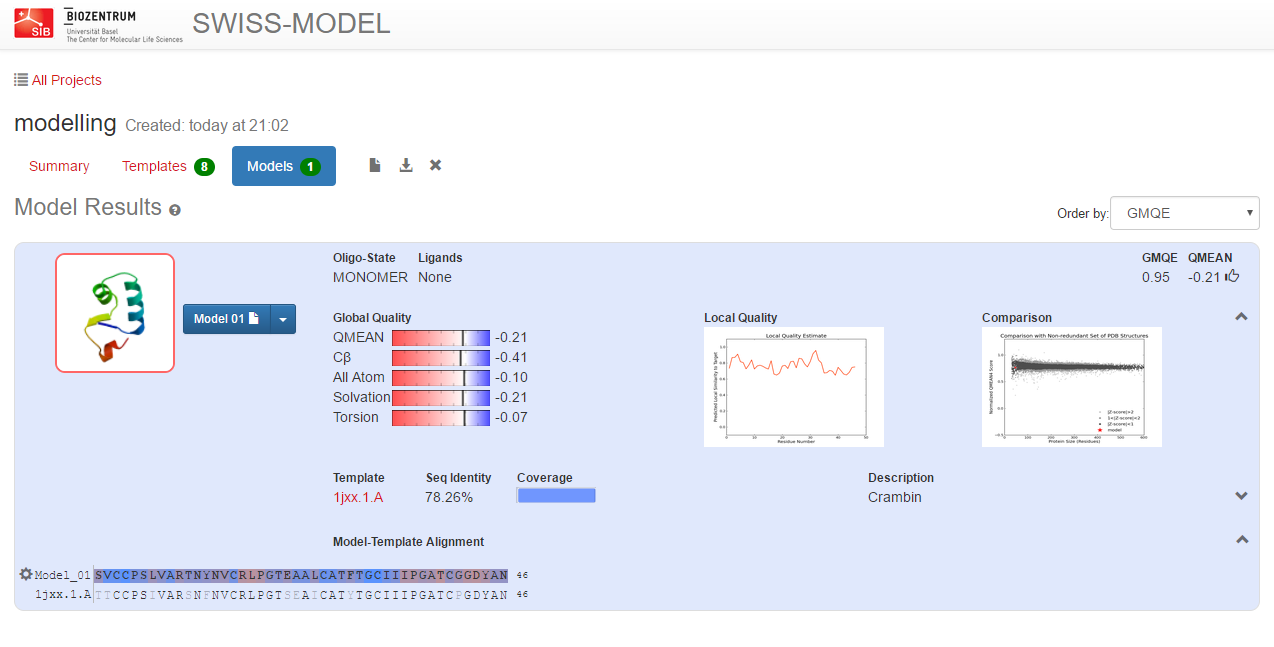
The server will alignment of target sequence and template structure(s), build a model and evaluate it. These steps require specialized software and integrate up-to-date protein sequence and structure databases. Each of the above steps can be repeated interactively until a satisfying modelling result is achieved.
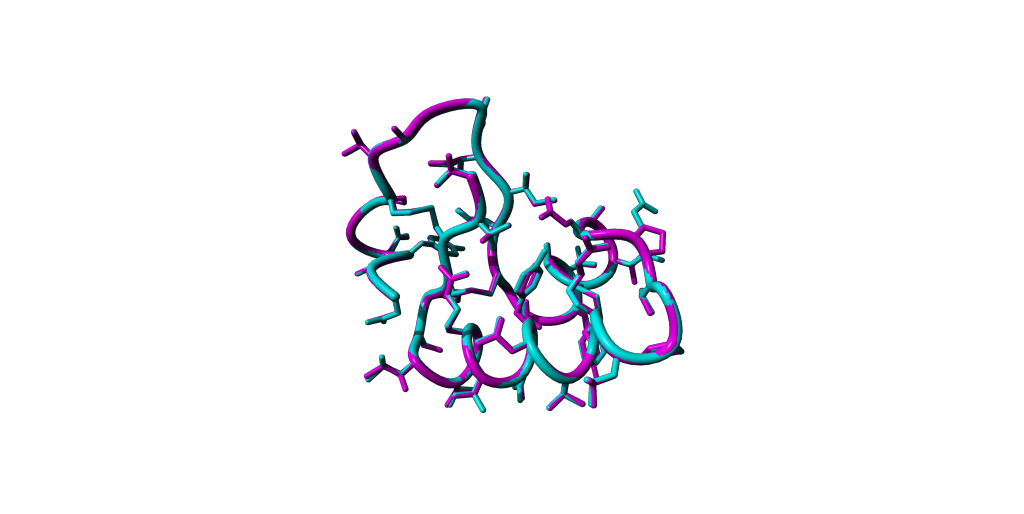
You can download the modelled structure as PDB format and load it into Yasara. Please perform a structural superposition with your reference e.g. 1CRN and try to detect the differences through manipulating the visualisations.