Archive for Module 3
Go to parent Basic bioinformatics concepts, databases and tools#Exercises_during_the_training
Version 5 Nov 2011 - JJ
Version 4 June 2011 - JJ
Version 3 Feb 2011 - JJ
Version 2 - Joachim Jacob
Version 1 - Guy Bottu
Contents
Additional exercises
Searching a sequence for domains in Pfam
Pfam is a protein domain database. Although it has no annotation of its own, it is very performant for finding protein domains. The HMM for each domain can be downloaded and a list of sequences on your computer (in fasta format) can be searched using this HMM through the HMMer program.
FYI: Pfam is composed of two parts: 1. Pfam-A, composed of Hidden Markov Models (HMM), made and searched using the HMMER suite of Sean Eddy. 2. Pfam-B, composed of alignments, nonredundant with Pfam-A, made using BLAST and the ADDA clustering software, searched by BLAST against the consensus sequences.
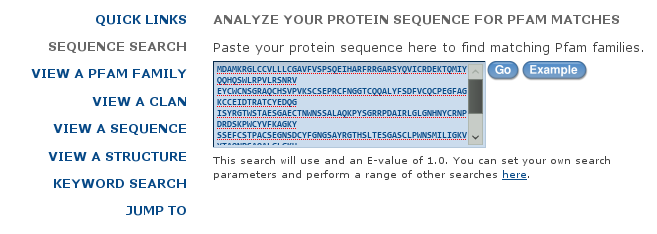
Go to http://pfam.sanger.ac.uk and follow the link "SEARCH". Copy-and-paste the content of TPA_HUMAN.fasta into the "Sequence" box and "Submit".
You will notice that the output contains information about which range of the sequence matches which range of the HMM. For EGF you might see a 31 in red, indicating that the match with the HMM was only partial. You can click on "Show" to see the ranges of the sequence that match the motif. You can obtain more information about the motifs.
Click e.g. on "Kringle". You can get some information about the HMM by clicking on "Curation & models". You can see the "seed alignment" that was used to generate the HMM as it is in Pfam by clicking "Alignments", setting "Viewer" to "Pfam viewer" and clicking "View".
Note: HMMer can be installed and run locally. See http://hmmer.janelia.org/. Besides searching your sequences for a domain (with a HMM), you can create a HMM yourself from an alignment using this software.
Sensitivity and selectivity: calculation example
Recall the GalNAc prediction from above. Calculate the sensitivity and selectivity of both tools on the example sequence (if the database represents the 'truth').
Name: sp_P32781_A Length: 87
MQLLRCFSIFSVIASVLAQELTTICEQIPSPTLESTPYSLSTTTILANGKAMQGVFEYYKSVTFVSNCGSHPSTTSKGSPINTQYVF
NetOGlyc __________________...TT......S.T...T.....TTT.............................TT.......T....
OGPET ......................T......S.T......S..T..................S.T..S........T............
OGLYCBASE .....................TT......S.T......S.....................S.T..S........T............
|
|---|
|
|
Searching RNA domains in Rfam
Rfam is a database of RNA motifs, defined so as to take account as well of conserved positions (bases) as secondary structure (base pairs). It is useful for predicting the secondary structure of an RNA molecule by homology with known structures and for searching noncoding RNA genes in genomes. It contains Covariance Models (CM), which are a variety of Stochastic generative Grammars (level Context-Free grammar) and is made using the INFERNAL suite of Sean Eddy.
We will as example submit a sequence of Salmonella typhimurium genomic DNA containing a 4 tRNA gene operon and see if we can find the localization of the tRNA
genes.
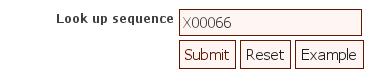
Go to http://rfam.sanger.ac.uk and follow the link "SEARCH". Copy-and-paste the content of X00066.fasta into the "Sequence" box and "Submit". Since the computations are quite elaborate it could happen that we must wait too long, especially if many submit at the same time. In that case you can just type x00066 into the "Look up sequence" box, the interface will send you a precomputed result instead of performing the search.
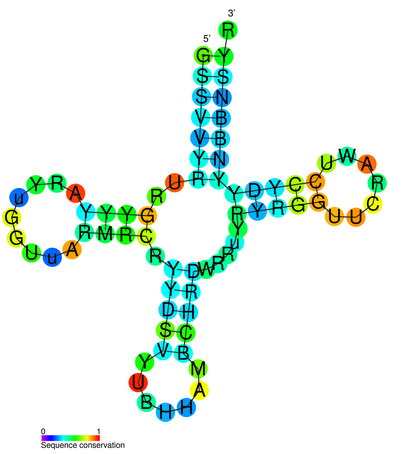
You will see that you get indeed 4 hits against the "tRNA" motif1. You can obtain more information about the motifs. Click "tRNA". Rfam itself contains a rather minimal annotation, but for some entries like this one it obtains information from (would you believe?) Wikipedia.
You can get some information about the CM (covariance model) by clicking on "Curation". You can get information about conserved secondary structure by clicking on Secondary structure". You can see the "seed alignment" that was used to generate the CM as it is in Rfam by clicking "Alignments", setting "Viewer" to "HTML" and clicking "View".
Note 1:to win time the interface at the Sanger Center first performs a WU-BLAST search against the sequence fragments from Rfam and then performs the INFERNAL search against this "filtered" data set. The INFERNAL search takes a lot of time because it involves dynamic programming in 3 instead of in 2 dimensions.
Note 2: you can download the INFERNAL suite and perform the search yourself without filter or with a more sensitive filter based on HMM's rather than on BLAST local similarities. You can also search a sequence only against selected models instead of against the complete Rfam.
More gene prediction tools
GeneMark for eukaryotes
Another gene searching tool is GeneMark.HMM. It has been developed by Mark Borodovsky at the Georgia Institute of Technology and is commercialized by Gene Probe inc. GeneMark.HMM exists in a version for eukaryotes and a version for prokaryotes. Contrary to GENSCAN GeneMark.HMM eukaryotic does not try to find signals. GeneMark is interesting because it offers HMM models for a large variety of organisms.
Go to http://www.geneprobe.net and follow the links "About Us", "Georgia Institute of Technology", "GeneMark.hmm", "GeneMark.hmm for eukaryotes".
Upload X02419.fasta, set "Species" to "H.sapiens" and "Start GeneMark.hmm".
You will note that GeneMark did miss the last exon. This shows that gene prediction software is still not perfectly reliable. Do note that there exists a Genemark.HMM eukaryotic version 3 that does work fine on this gene ; it is not available on the public server but you can install it locally after you have obtained a licence (free for academics).
GeneMark for prokaryotes
GeneMark is one of the few gene searching tools that pays attention to prokaryotes. GeneMark.HMM prokaryotic is accompanied by a huge number of models for
different eubacteria and archaebacteria. For some species it also models the ribosome binding site (RBS). In case the organism you are working on (or a closely related
one) is not in the list, GeneMark has so-called "heuristic" models, based on genetic code and %GC. We will take as example the EMBL entry with ID/AC V00307. It contains the ompA gene (coding for a porin) and an ORF that has only much later after the sequence had been entered in EMBL/GenBank been shown to code for a component of the S.O.S.
system.
Go back to the page where you can choose between different versions of GeneMark.HMM and go to "GeneMark.hmm for prokaryotes". Do note how many
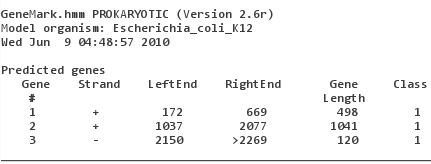
models for different species are available! Upload V00307.fasta, make sure that "Species" is set to "Escherichia_coli_K12" and "Start GeneMark.hmm".
You will note that GeneMark did find the two genes (and made a probably false-positive prediction of a reading frame running over the 3'-end). "Class 1" in the
output refers to the fact that for most organisms GeneMark makes a difference between "typical genes" (class 1) and "atypical genes" (class 2).
De novo prediction of mucin-type glycosylation in protein sequences
Let's do this protein sequence analysis to predict post-translational modification.
Many proteins require glycosylation for proper functioning. Techniques exist to determine glycosylation in the wetlab: but prior of going to the wetlab, you can predict glycosylation sites in sequences. Tools that do so try to mimick how the glycosylation machinery scan the sequence to detect the signals. In practice, it comes down again to patterns, motifs or machine learning techniques (such as Hidden Markov Models). Try to solve following task.
Can you tell with some confidence which sites are O-GalNAc glycosylated in the yeast agglutinin-binding subunit P32781? Use at least two tools and combine the results. (Try to do it by yourself before clicking 'Show'!)>sp|P32781|AGA2_YEAST A-agglutinin-binding subunit OS=Saccharomyces cerevisiae GN=AGA2 PE=1 SV=1 MQLLRCFSIFSVIASVLAQELTTICEQIPSPTLESTPYSLSTTTILANGKAMQGVFEYYKSVTFVSNCGSHPSTTSKGSPINTQYVF |
|---|
| * For all our protein prediction tools, we look on Expasy http://expasy.org/tools/. |
| * Let's use our browser as we should: press ctrl+f, a search box should appear and we type GalNAc to search on the page. |
| * Two tools are available at Expasy: http://www.cbs.dtu.dk/services/NetOGlyc/ and http://ogpet.utep.edu/OGPET/. Let's try both of them! (perhaps searching the literature may unveil more prediction tools): |
| * A tip: open both tools in a new tab in your browser by clicking with the middle button on your mouse. |
| Since so many tools are available, you should always take some time to read through the manual of both tools. |
| * Let's get the sequence of P32781 and download the sequence in fasta in a file on your computer (P32781.fas). |
| * Enter the sequence in respective input boxes of both tools. |
| * Combine the results you obtain to compare them. you could do this in an simple editor such as wordpad. |
Name: sp_P32781_A Length: 87
MQLLRCFSIFSVIASVLAQELTTICEQIPSPTLESTPYSLSTTTILANGKAMQGVFEYYKSVTFVSNCGSHPSTTSKGSPINTQYVF
NetOGlyc __________________...TT......S.T...T.....TTT.............................TT.......T....
OGPET ......................T......S.T......S..T..................S.T..S........T............
|
| * We see that some sites correspond, and others do not. It is fair to say that we have more confidence in overlapping sites. |
| * But perhaps we can find a database on experimental glycosylation results? Google for it to find at least one. Note that several exist: although they all surely do their best, some interfaces are just to inadequate. Here is one I found: http://www.cbs.dtu.dk/databases/OGLYCBASE/Oglyc.base.html. |
| * Combine experimental evidence with the predictions: which tool is most correct? |
Name: sp_P32781_A Length: 87
MQLLRCFSIFSVIASVLAQELTTICEQIPSPTLESTPYSLSTTTILANGKAMQGVFEYYKSVTFVSNCGSHPSTTSKGSPINTQYVF
NetOGlyc __________________...TT......S.T...T.....TTT.............................TT.......T....
OGPET ......................T......S.T......S..T..................S.T..S........T............
OGLYCBASE .....................TT......S.T......S.....................S.T..S........T............
|
| From the 9 experimentally confirmed results, NetOGlyc misses 1, but reports 4 not present in the database. In contrast, while OGPET misses one, it erronously reports also one. Perhaps on this sequence, OGPET performs better. |
Go to parent Basic bioinformatics concepts, databases and tools#Exercises_during_the_training