Variation data
Go to parent Basic bioinformatics concepts, databases and tools#Exercises_during_the_training
Contents
Sequence variation information
*Ensembl
*SNPs in human F9
We come back to the search for the human F9 (Coagulation factor IX Precursor) gene in Ensembl since Ensembl contains a vast amount of sequence variation information. First to the Gene page.
Click on Sequence in the left menu.

You now see the genomic sequence of F9 on the gene page. Exons are highlighted in red.
Click Configure this page in the left menu to open the configure page window:
- For Show variations select Yes and show links
- For Number of base pairs per row select 90 bps
- For Line numbering select Relative to this sequence

Close the window to allow the changes to take effect.
Now all SNPs and small variations are highlighted on the sequence following a colour code that represents the consequences of the varations and is explained at the top. At the right you see links that you can click to find more info on that specific variation. The IDs that start with "rs" are from dbSNP (check the overview of all variation databases that were used in Ensembl's Variation pages).
Click the rs number of the variant on position 722.

This takes you to the corresponding Variation page.
| What kind of variant is this ? |
|---|
 It's a SNP (red box). |
| What are the alleles of this variant ? |
|---|
 It's a SNP with alleles G/A (green box). |
| In which part of the gene is the variant located ? |
|---|
 It's located in an intron (blue box). |
| Was it found in the 1000 Genomes project ? |
|---|
 Yes, according to the evidence codes (purple box). Click the i next to Evidence status to see the meaning of the codes. You see here that this variant has multiple independent dbSNP submissions, i.e. submissions from different submitters or different samples ! |
More detailed information is available when you click one of the icons. For instance click the Population genetics icon.
| In which populations do you find the alternative allele ? |
|---|
 The alternative allele is found in the African and American population (albeit at very low frequencies), not in the European or Asian populations. |
For this variant, there's no phenotypic info available but for some of the variations, their phenotypic implications are known and documented in Ensembl.
*Somatic mutations in human F9
Go back to the Gene page of F9 and click the COSMIC ID (COSM385264) of the variant on position 635. COSMIC is a database of somatic mutations found in human cancers (check the overview of all variant databases used by Ensembl.
| What type of cancer is this variation implicated in ? |
|---|
| Click the Phenotype data icon. The variation is implicated in lung tumours. |
| In which transcripts is this mutation found ? |
|---|
| Click Genes and regulation in the left menu. The variation is found in all three F9 transcripts. |
| What is the ancestral allele ? Is it conserved in primates ? |
|---|
| Click Phylogenetic context in the left menu. The ancestral allele is G. A region containing the somatic SNV (placed in the centre) and its flanking sequence is displayed. The G allele is conserved in all primates.
 |
*All diseases F9 is linked to
Go back to the Gene page of F9.
| Get an overview of all diseases F9 and its variants are implicated in |
|---|
| Click Phenotype in the left menu.
 On the Phenotype page phenotypes that have been associated with the F9 gene as well as with variants associated with the F9 gene are shown.
 |
*Overview of all variantions in F9
| How many coding sequence variants are known for F9 ? |
|---|
| Another way to view the variations is clicking the Variation table in the left menu to display an overview of all variations in F9.
 In the Variation table all variants of the F9 gene are shown.
 Now you only see these 5000 CDS variants in the table. Clicking the ID of a variant redirects you to the corresponding Variation page. |
*Large variations in F9
All information of dbVar, NCBI's database of large(r) variations, is included in Ensembl.
Large variations can be viewed by clicking Structural variants in the left menu. This opens a graph and a table of known structural variations in the F9 gene sequence. Remember that CNV stands for copy number variants.
| Does F9 have copy number variants ? |
|---|
| Yes, smaller and larger CNVs have been annotated for this gene, as indicated in the SV smaller variants and SV larger variants track. Details are given in the table below the graphical display. |
*Variations in a transcript
Go to the transcript page of F9-001, its longest transcript.
Here also you can view the variations in the sequence.
- Click Exons under the Sequence menu item in the left menu to show the sequences of the exons. Remember that UTRs are displayed in purple, exons in black, introns in blue and upstream and downstream sequences in green.
- Use configure this page to show variations as we did on the gene page (it might be a good idea to select Show full intronic sequences if you want to view variations in introns).
- To export the view with the variations highlighted click Download Sequence and select RTF format.
If you only want to view variations in the CDS, click cDNA under the Sequence menu item in the left menu. Variations are displayed by default.
Again there are also other ways to view variation info.
| The R on the third position of the protein encoded by this transcript is sometimes mutated into another amino acid. Which one and what is the dbSNP ID of this missense mutation ? |
|---|
| On the transcript page, go to the left menu and click Variants in the Protein information section. This opens the variations table on the transcript page. When you scroll down you see that there is indeed a missense variant on position 3 of the sequence that transforms the R into a H. A missense variant is a sequence variant, that changes one or more bases, resulting in a different amino acid sequence but where the length is preserved. The dbSNP ID of this variant is rs148060786. |
On the Locations page you can visualize variations by adding Variation tracks.
*Variant effect predictor
Exercise developed by EBI
Resequencing the genomic region of the human CFTR (cystic fibrosis transmembrane conductance regulator gene) has revealed the following variants (alleles defined in the forward strand):
- G/A at 7:117,171,039
- T/C at 7:117,171,092
- T/C at 7:117,171,122
The Variant Effect Predictor tool allows to predict the functional consequences of these variants.
| Predict if these mutations will change proteins encoded by any of the genes in this region? Which gene? |
|---|
Go to the Ensembl home page and click the link Tools at the top of the page. Click Variant Effect Predictor and enter the three variants as below:
7 117171039 117171039 G/A 7 117171092 117171092 T/C 7 117171122 117171122 T/C
You will get a table with the consequence terms from the Sequence Ontology project (http://www.sequenceontology.org/) (i.e. synonymous, missense, downstream, intronic, 5’ UTR, 3’ UTR, etc) for the listed SNPs. These are all intronic variants of the gene suppression of tumorigenicity 7 (click the Ensembl ID to see the name of the gene). |
*NCBI's variation resources
*Variant reporter
We sequenced blood samples from various patients with a disease and their family members. After analyzing the reads we obtained a list of sequence variants that are specific for the patients. We want to check if the variants that we find are novel.
To see if variants are already known you need to use NCBI's variant reporter tool. We are going to do the analysis on three variants but you can submit as many variants as you want.
The variants have to be submitted in a certain format but many formats are supported. Check the help files to see which other formats are supported. We will use the VCF format since this is typically generated by NGS workflows. It's a tab-delimited text file, you can check out the VCF specification to see the details of this format.
Download the variants file in VCF format.
| Check if the mutation is already known. |
|---|
* Go to the variant reporter tool and select the organism to work in.
 As you can see in the results all three variants are already known and are present in dbSNP. |
Feel free to check out the dbSNP records of these SNPs.
Genomic Variation Server
GVS provides easy access to variation data from dbSNP, HAPMAP and other resources. You can define a location or region of the human genome to search in and GVS will give you access to all available variation info in that region. As an example, we'll take a look at the variation in BRCA2.Go to the GVS website.
| Using BRCA2 as a keyword how are we going to search the database ? |
|---|
| Since BRCA2 is a gene name you have to do a search based on gene name. |
| Perform the search |
|---|
*Type BRCA2 in the search box. If you provide a gene name GVS will search in the complete gene sequence, introns and UTRs included if they are annotated. Since BRCA2 is a well annotated gene we do not want to include regions up- or downstream of the gene. If UTRs are not annotated or you want to search for variations in the promoter of the gene you might want to include up- and downstream regions.
 |
This returns an overview of all data sets containing data on variants in BRCA2.
| Obtain a summary of SNPs in the BRCA2 gene that occur in at least 48% of the Japanese (HAPMAP-JPT)? |
|---|
The Japanese are represented in the HAPMAP JPT study. You can get more info about a population by clicking its name.
Scroll down to set the parameters of the search: you can get more info about a parameter by clicking its name: this will take you to the GVS manual.
 |
Now you see an overview of the SNPs in the BRCA2 gene that occur in more than 48% of the Japanese population. As you can see, these are all intron variants and one synonymous substitution (different codon encoding same amino acid). It is expected that variants that occur at such high frequencies have no impact on the BRCA2 protein sequence.

| Obtain a linkage disequilibrium scores for these SNPs |
|---|
| Go back to the search site and leave the parameters as they are. Click display linkage disequilibrium. |
For each pair of SNPs the r2 score represents the number of cases that both SNPs occur in the same person (ranging from 0 to 1). The idea behind this is that if two individuals share the same variant, we would also expect that they share not just that variant but also the surrounding chromosomal region.
| Check out what happens when you display tag SNP |
|---|
| Go back to the search site and leave the parameters as they are. Click display tag snp.
 |
This groups SNPs based on r2 values. This is useful for the development of a minimal set of SNPs for genotyping similar populations (by selecting one SNP from each bin). The Tag SNPs are those for which the pairwise-r2 values exceed the r2 Threshold. The Other SNPs are those for which the pairwise-r2 values are less than the threshold. It is better to choose a SNP from Tag SNPs to represent the bin.
Database of genomic variants
Go to the query tool of the database of genomic variants. The database is a set of inter-related tables containing all the data from the studies included in DGV. You can search and filter the data in different ways, e.g.
- data that come from a particular studyy
- variants of a certain type e.g. copy number variations
- sample size e.g. variants coming from large population studies
- ...
You can set multiple filters at the same time.
| Find all variants located on the Y chromosome in assembly hg19 |
|---|
* Select the chromosome filter and set it to Y
 Go to the Variants tab. |
This returns over 6000 variants on chromosome Y (found by mapping to assembly version hg19) coming from different studies. As you can see at the top right of the list, you can save the output in various formats.
| What is shown on the Study tab ? |
|---|
| The Stud tab shows studies which have identified variants on the Y chromosome. |
| What is shown on the Platform tab ? |
|---|
| The platforms used in the studies which have identified variants on the Y chromosome. Most of the studies were done by Next Gen Sequencing but not all of them. |
OMIM
Exercise obtained from OpenHelix.
Exercise 1: the human RANKL gene
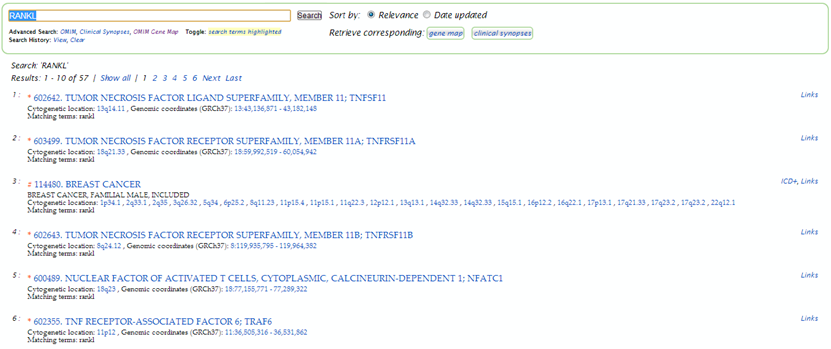
I would like to know if there are any phenotypes associated with the human RANKL gene, and whether this association is due to variation in the gene. Do a basic OMIM search for the gene RANKL, also known as TNFSF11 (see slides).
| What is the genomic location of the gene ? |
|---|
| A basic search on rankl generates a list of 57 results.
|
| What disease is associated with the gene ? |
|---|
| In the Table of Contents (see slides), click the Table View link found under Allelic Variants. This opens a table of allelic variants associated with osteopetrosis. |
| Can you locate other reports that offer information on phenotypes that might be similar to yours ? |
|---|
| Return to the gene record. In the Gene Phenotype Relationships table click the link to the phenotype record. Click the Phenotypic Series link to see information on phenotypes that might be similar but that occur in other genomic locations. |
Exercise 2: myopathy
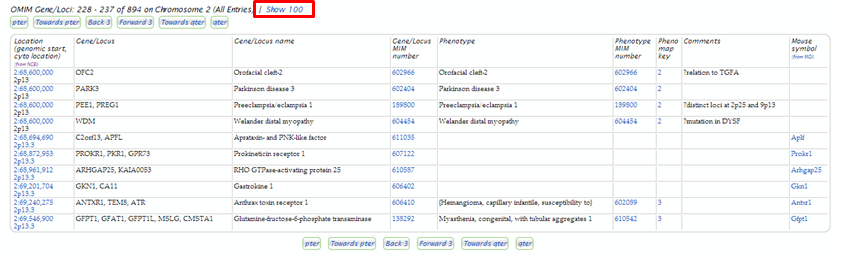
Imagine that you have a patient displaying evidence of myopathy that has been linked to the chromosomal location 2p13. Conduct an OMIM Gene Map search for the area (see slides). On the search results page assess phenotypes in the region that may be the cause.
| How was this phenotype placed on the map ? |
|---|
| Mouse over the 2 in the Pheno map key column to determine how this phenotype was placed on the map. |
| Are there more genes in this region linked to myopathy ? |
|---|
| Scroll or use your browser’s Find function to locate the next gene or phenotype description that includes the term myopathy</b<. You will find the TIA1 and the Dysferlin gene associated with a myopathy phenotype. Dysferlin is also associated with 2 muscular dystrophy phenotypes. |
To learn more about the Dysferlin gene, under the Gene/Locus MIM number column, click the <b>606768 link to open the gene report.
| How many exons does the dysferlin gene contain ? |
|---|
| In the Table of Contents and click Gene Structure. |
| What kind of mutation of the dysferlin gene is associated with myopathy ? |
|---|
| In the Gene Phenotype Relationships table click OMIM ID of the myopathy phenotype to go to the phenotype record. In the Table of Contents and click "Molecular Genetics". |
Exercise 3: rheumatoid arthritis
| How can you perform the following basic search in OMIM: "rheumatoid OR arthritis" ? |
|---|
| Using the OMIM basic search box, enter the text: "rheumatoid arthritis". Then click “Search”. |
| How can you perform the following basic search in OMIM: "arthritis NOT rheumatoid" ? |
|---|
| Using the OMIM basic search box, enter the text: "-rheumatoid arthritis". Then click “Search”. |
| Why was BLAU SYNDROME returned by the search ? It doesn't contain the word "arthritis". |
|---|
| Click on the link to go to the phenotype record. Examine the alternative titles area: arthritis is found in one of the alternative titles. |
Analyzing NGS data for variant analysis
Go to our NGS wiki page for the introductory tutorial on NGS data analysis (checking/improving the quality of your data, mapping the reads, obtaining the tools/data) and the tutorial on variant analysis.