Mappers
This page lists 'mappers', also called 'aligners', of next-generation sequencing data. These programs map/align NGS data to a reference genome, which usually needs to be prepared or indexed prior by that mapper. As reference genome, the reference of the species is preferably taken, but also other species' reference genomes can be used to map to, although with much lower quality results.
Many mappers exist and are being developed, due to the specific computational problem: hundred thousands to millions of relatively short reads (30 to 500 bp) need to be mapped to the reference genome, which is 7 to 8 times an order of magnitude larger. This task needs to be performed with taking into account variations of the reads: insertions, deletions, mutations, orientation (in case of paired-end reads), splice-junctions (in case of mapping RNAseq data),...
![]() A selection of our favorite mappers is presented in the DNAseq_toolbox
A selection of our favorite mappers is presented in the DNAseq_toolbox
Contents
List of mappers
See also
- http://wwwdev.ebi.ac.uk/fg/hts_mappers/
- RNA-seq mappers: http://www.rna-seqblog.com/data-analysis/splicing-junction/rna-seq-alignment-tools/
Benchmarking mappers
Benchmark tests can asses computational requirements, as well as biological validity of the mapping results. Below we focus on the accuracy of the mappers, which is extremely important in some secondary analyses such as variant calling.
Methods
Simulating reads
To benchmark mappers, these methods start from a reference genome of interest for simulating a sequencing run, resulting in simulated reads. Based on the performance of the mapper on these reads against the reference, accuracy statistics are collected. The model generating the reads has to account for many known and yet unknown biases and errors influencing read count and quality, depending on the mimicked platform and the genome generating the reads from.
See Read simulation page for read simulators.
Simulating a reference genome
ARDEN ([5]) does not simulate reads, but alters the reference genome to an artificial genome, following some rules, such that none of the reads aligns perfectly any more to the reference. ARDEN compares the mapping to the reference genome with the mapping to this artificial genome, from which it calculates sensitivity and specificity.
Analysis of real reads that align imperfectly
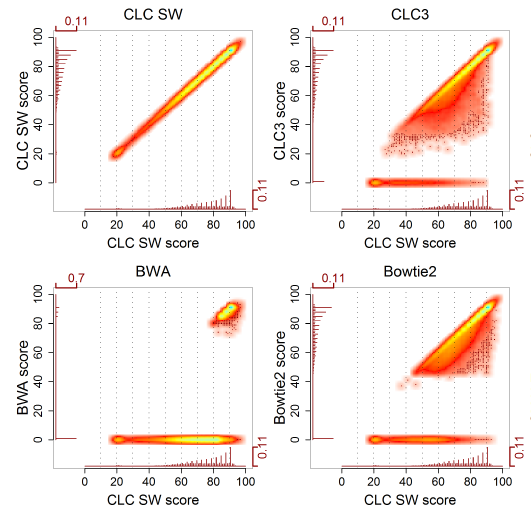
CLC Bio has used a different approach to benchmark mappers ([6]). The rationale is this: the best mapping we can perform is the Smith-Waterman (SW) algorithm. Secondly, reads mapping perfectly do not provide information on accuracy. Hence, they have analysed a subset of real reads which do not align perfectly, aligned them with SW and with the mapper of interest, and compared the results. Typically, many reads were mapped optimally, a large subset were mapped suboptimally, and some reads were unmapped. The results are displayed in a scatter plot, comparing the optimal score of an alignment (by SW) with the score obtained by the heuristic mapper.