Getting and manipulating genome tracks in Galaxy
Go to back to Galaxy beginner's tutorial#Galaxy 'DNA' workshop exercises
In this tutorial, we will fetch data and tracks from 'third-party' data sources, such as the UCSC Genome Browser or Biomart. You can find the tools to import these data under the section 'Get Data'.
Contents
Getting data from UCSC
You can get genomic information from UCSC. We will fetch the refseq exon coordinates of chromosome 21 of the human genome build 37 (hg19). Because we want to know where the SNPs from the previous exercise are located in exons (for example).
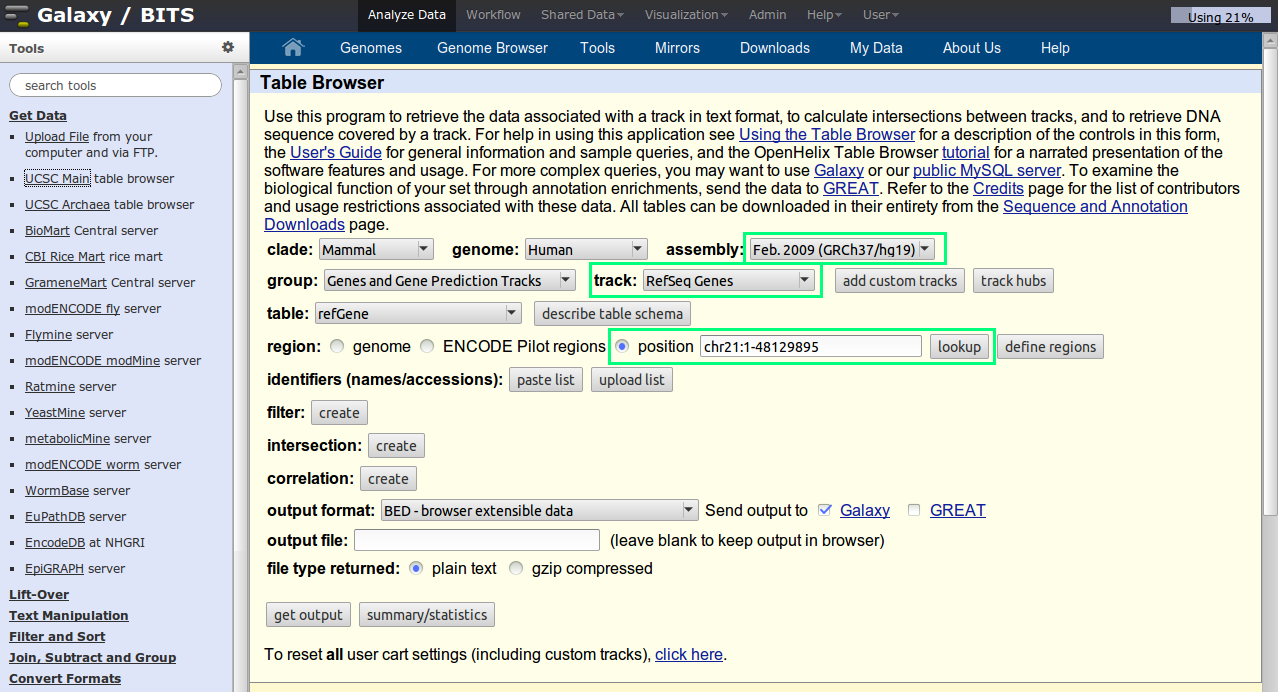
Click on UCSC Main table browser in the 'Get Data' section. The default UCSC table browser will appear in the middle pane. Set the correct parameters in the UCSC page. Select the right species, and genome build. Select the refseq gene track.
One small tip: to set the chr21, enter chr21 in the box and click op lookup. The coordinates are automatically added.
Your final settings should look like this:
Note that the output is set to be send to Galaxy.
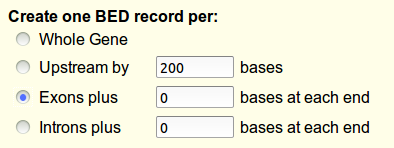
In the next screen, you can select which coordinates you want to send: we choose the exon boundaries, and click 'Send to Galaxy'.
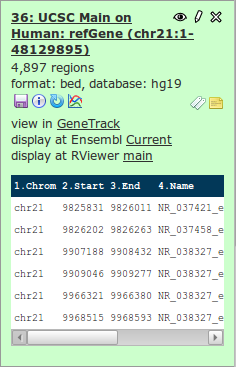
A new dataset is being generated, with genome coordinates from UCSC. Look at the data and notice the data type being used.
Galaxy tools to calculate with genomic coordinates
The dataset just fetched from UCSC is in BED format (a type of tabular text format). The BED format is also a format - just like interval format seen before - which stores coordinates of genomic annotations, optionally enriched with more information. See a description of the BED format here.
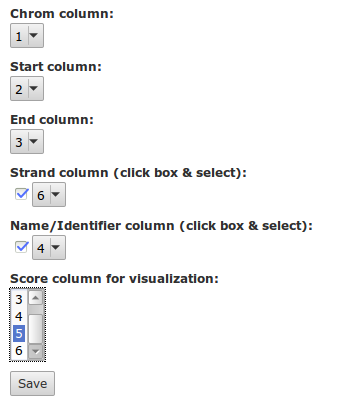
The first three columns of BED are obligatory. Galaxy knows what is in these columns when using BED: you can preview them. The fourth and sixth column Galaxy has correctly identified as being the name and strand respectively. But we have also a column with the score (column 5), which Galaxy does not know about. Edit the attributes of the dataset.
Change the name of this dataset to Exons chr21 hg19. A good practice is to copy the original name to the info box in Galaxy.
Once we have such tracks of genomic annotations, we can do calculations with them. Galaxy has a nice set of tools under the section Operate on genomic intervals.
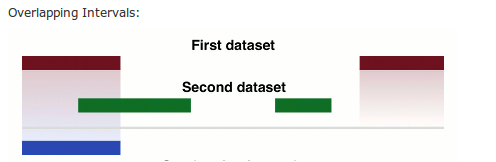
We can filter our SNP list for now those located in exons. We choose 'Intersect the intervals of two datasets'. This tool has a nice help section at the bottom.
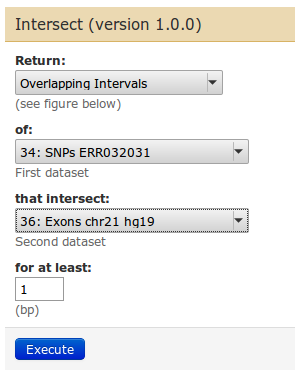
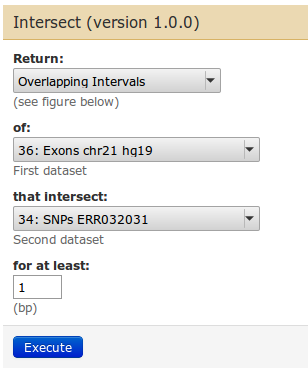
To get to our goal, we choose parameters as follows:
| Task: filter now the exon list for exons that only contain SNPs. |
|---|
|
From the data set preview we see that of 4,897 exons, 1,619 exons contain SNPs. Of 13,050 SNPs, 5,316 are located in exons.
Visualize the two tracks next to each other in Galaxy
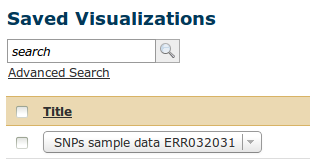
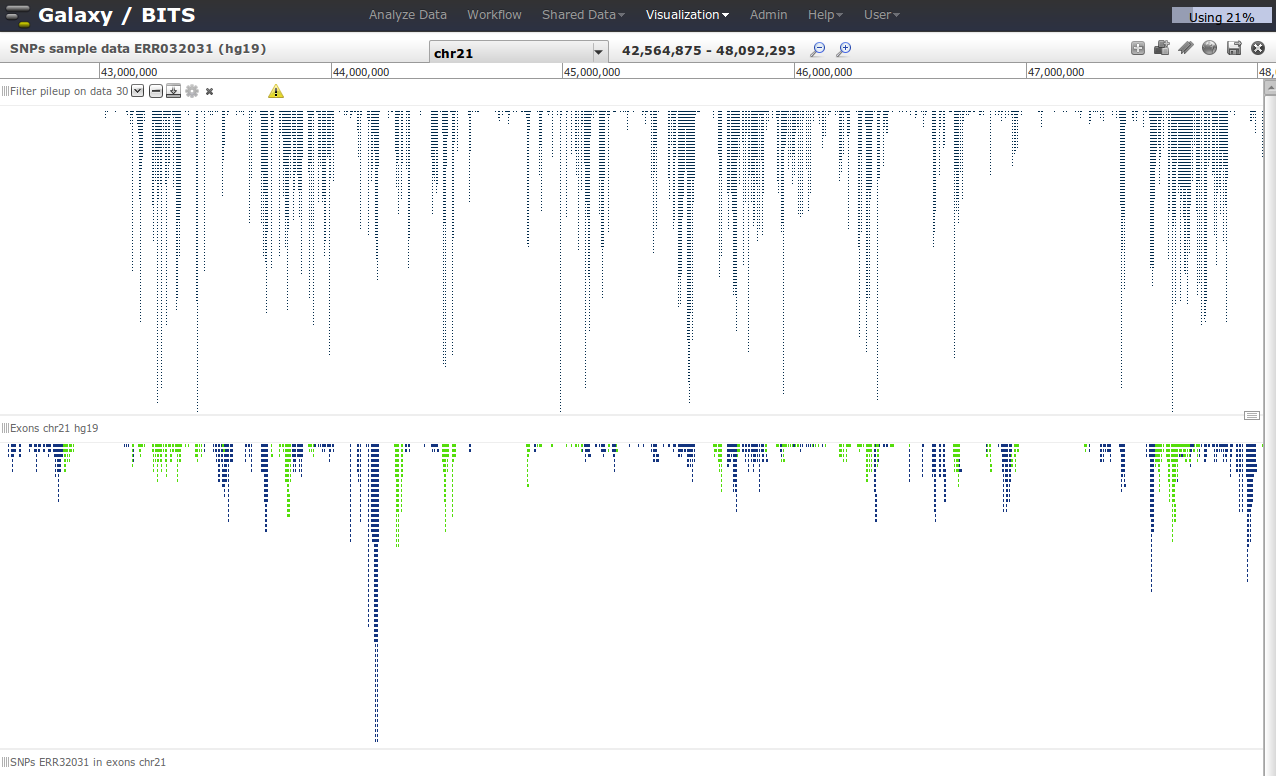
First we go via the top menu 'Visualisation' to our 'Saved visualisation' called 'SNPs sample data ERR032031'.
If the visualisation is loaded, we can add tracks from our current history to the visualisation by clicking in the right top corner 'Add tracks'.
The action is going on on chromosome 21, so do not forget to select that region. Also, do not forget to save regularly your view by clicking in the right upper corner ![]() .
.
Visualizing data and deciding on what to do next is crucial in designing your analyses. You can quickly shift between the analyze data modus and visualisation via the top menu bar.
Counting the SNPs per exon
See if you can do this yourself. One hint: a SNPs is one position
| Show me where to start! |
|---|
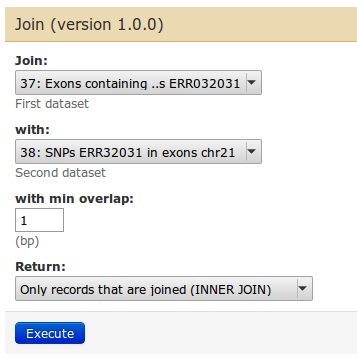
| Use 'Join the intervals of two datasets side by side'. The idea is to get every exon line replicated as many times as there are SNPs in that exon. The 'Join' tool can do that. Afterwards, we will count how many times each exons occurs. |
| Show me the steps! |
|---|
| |
|

|
We count on the exon name, which is in columns 4. So choose 'c4' 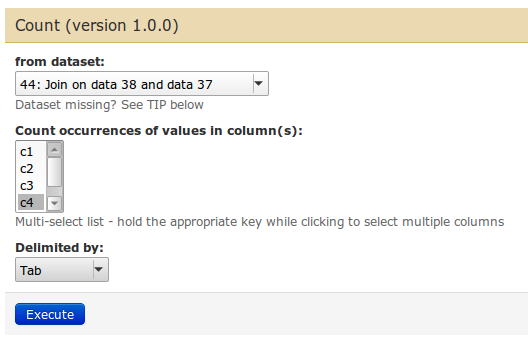
|

|
|
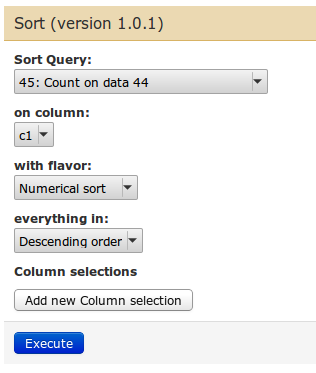
| We have our list of exons with most SNPs! You can combine them |
With this exercise we end the 'DNA seq' tutorial. Have fun with Galaxy!
Go to back to Galaxy beginner's tutorial#Galaxy 'DNA' workshop exercises