DNA sequence analysis
Go to parent Basic bioinformatics concepts, databases and tools#Exercises_during_the_training
Contents
Gene regulation via Ensembl
At lot of sequence analysis has already been done for you in specialized sequence databases such as Ensembl. Go to the region from bp 52,000,000 to 52,200,000 on human chromosome 4.

- Zoom in on the SGCB transcript, including a bit of flanking sequence on both sides.
- In the middle graphic select the Drag/Select
 button and draw with your mouse a box around the SGCB transcript. If you don't see the button, it means that Drag/Select has already been activated.
button and draw with your mouse a box around the SGCB transcript. If you don't see the button, it means that Drag/Select has already been activated. - Click Jump to region in the pop-up menu

CpG islands are genomic regions that contain a high frequency of CG dinucleotides and are often located near the promoters of mammalian genes. There are tools that predict potential CpG islands based on nucleotide composition. These predictions can be visualized in Ensembl.
| Is there a CpG island located at the 5’ end of the SGCB transcript ? |
|---|

 When you scroll down at the most detailed display, you see that there is indeed a CpG island located at the 5’ end of the SGCB transcript.
 |
Possible transcription start sites can be predicted using the Eponine tool. Eponine predictions are one of the tracks available in Ensembl.
| Is there a transcription start site predicted by Eponine annotated for the SGCB transcript? |
|---|
When you scroll down at the most detailed display, you see that Eponine indeed predicts a transcription start site almost exactly at the same location where the Ensembl SGCB transcript starts.
 |
An important mechanism of gene regulation is the accessibility of the DNA. For human and mouse Ensembl provides several tracks for visualizing the status of the chromatin, based on data from the ENCODE project.
This project aims to experimentally characterize all functional elements in the human and mouse genome: TFBS, TSS, open chromatin regions, non-protein-coding transcripts... The project obtains this information among others from large scale ChIP-Seq or DNA-Seq experiments like the sequencing of DNA regions that were cut by DNase to identify closed and open chromatin regions in the DNA.
| Is the chromatin in the vicinity of the SCGB gene open or closed in H1ESC cells ? |
|---|

 When you scroll down at the most detailed display, you see that the DNase method identifies an open chromatin region that overlaps with the CpG island and the predicted TSS.
 |
Similarly you can also define TSS as the regions that bind to the RNA polymerase. Encode performed ChIP-Seq experiments on various cell type samples to identify DNA regions that bind to RNA polymerase.
| Does the experimentally determined TSS correspond to the one that was predicted by Eponine ? |
|---|

 When you scroll down at the most detailed display, you see that the fragment that binds to PolII according to the ChIP-Seq experiment indeed overlaps with the predicted TSS.
 |
Finally, DNA accessibility can also be regulated by modification of histone proteins. DNA winds itself around these proteins and as such they regulate the accessibility. If the DNA is tightly wound aroound them, it is not accessible. If the DNA loosens from the histones it becomes accessible. how well the DNA can bind to the histones is determined by the structure of the histones: i.e. which modifications are on the surface of the histone proteins ? Methyl groups, acyl groups or other modifications all influence the binding of the DNA .
ENCODE performed ChIP-Seq experiments with antibodies that target modified histone proteins to identify the DNA regions that are bound to them.
| Is the regulatory region at the 5' end of the SCGB transcript bound to methylated H3K4 ? |
|---|

 When you scroll down at the most detailed display, you see that the region is bound by doubly and triple methylated H3 in embryonic stem cells.
 Methylation of the fourth amino acid residue (K) from the N-terminus of histone H3 is one of the most studied histone modifications, and with good reason: it’s tightly associated with the promoters of active genes. Like all lysine residues, H3K4 can be mono, di, or tri methylated. H3K4me3 is the strongest activator of expression. |
Finally, you can also search for binding sites of specific TFs in Ensembl, again based on ENCODE data.
| Which TFs have a binding site in the CpG island of the SGCB gene in embryonic stem cells ? |
|---|
 This track contains information from ENCODE ChIP-Seq experiments. As such it specifies the location of TFBS in the genome. You see a matrix with TFs in the rows and cell types in the columns.
 Hover your mouse over the H1ESC. A box will appear in which you can select to visualize all TFs for which data are available will on the track. Close the configure page
 You can see that TAF1, TAF7 and Yy1 have a binding site in the CpG island. TAF1 and TAF7 are subunits of TFIID, which binds to the TATA-box in the Transcription Start Region to position the RNA polymerase properly on the DNA. Yy1 represses transcription by directing histone modifying proteins to promoter regions. |
RNA processing via miRWalk
MiRWalk is a database that contains information on miRNAs from Human, Mouse and Rat. For each miRNA, it contains predicted as well as experimentally validated binding sites in their target genes.
In a mouse study, you find that the expression of TIMP3 influences the development of atherosclerosis. A decreased expression of TIMP3 stimulates inflammation of the arterial endothelium and triggers atherosclerosis.
You have searched for TFBS for TFs that might be the cause of the TIMP3 repression but to no avail. Maybe the expression of TIMP3 is regulated via other mechanisms, like miRNA induced degradation.
For decades, scientists have assumed that miRNAs bind to sequences in the 3'UTR of target genes.
| Is TIMP3 expression regulated by binding of miRNAs in 3'UTR ? |
|---|

 The results page contains
When you click the results for the 3'UTR, you indeed get a list of miRNAs that in theory could bind to the 3'UTR of TIMP3 . |
You spend a lot of time checking if these miRNA are the cause of the decreased TIMP3 expression in atherosclerosis but you find no link at all. Quite frustrated you decide to perform a final analysis with an open mind. Although everybody states that miRNAs target 3'UTRs, you decide to scan the CDS for potential targets.
| Do you find miRNA targets in the CDS of TIMP3 ? |
|---|
Fill in the form as follows and click SEARCH:
 miRWalk allows you to search in any component of a gene: 3'UTR, CDS, 5'UTR and promoter. You get a list of miRNAs that in theory could bind to the CDS of TIMP3, one of them is mir-172 (red) .
 |
You go back to the lab and find that atherosclerosis activates mir-172, which in its turn decreases the expression of TIMP3, thereby worsening the atherosclerosis.
![]() It is important to search miRNA binding sites within the complete sequence (promoter, 5'-UTR, CDS and 3'-UTR) of a gene !
It is important to search miRNA binding sites within the complete sequence (promoter, 5'-UTR, CDS and 3'-UTR) of a gene !
| Does mir-172 target other CDS ? |
|---|
For this we need to do the inverse analysis: start from a miRNA and predict which genes it targets. Fortunately, miRWalk can perform both analyses.
The miRNA-target search page is organized similar to the Gene-target search page. You select a species, provide an ID of the miRNA, select the regions you want to search in, set the options, select the algorithms you want to use for the predictions and click SEARCH.
 You get a list of CDS that can bind to mir-172, one of them is Pla2g6 (red).
 You go and search for a link between this gene and atherosclerosis in Pubmed and bingo: 10 publications containing the name of this protein and atherosclerosis. |
Find regulatory motifs using MotifLab
We have identified a number of genes that respond to DNA damage and stress, initiating cell-cycle arrest or triggering apoptosis. Download a list with the Ensembl IDs of the potential targets. The first thing you need to do is creating a new protocol (=analysis). I want to know which TFs might be responsible for the observed upregulation of these genes in response to stress.
| How to create a new protocol ? |
|---|
| Click the New Protocol button in the top menu
 |
If you want to repeat your analysis later, you can document everything you do now via the record function.
| How to record everything you do ? |
|---|
| Click the Record button in the top menu
 This will record your actions so it becomes easy to repeat the analysis on the same or other data. |
The first thing you have to do is to import the sequences of the promoters you want to analyze.
| How to import sequences ? |
|---|
You can import promoter sequences by just providing Gene IDs or Ensembl IDs. MotifLab is coupled to Ensembl and will fetch the appropriate promoter sequences automatically. Click the Add Sequences button in the top menu
 Paste the Ensembl IDs or import the file containing the IDs (red). Note that you can choose the exact sequence of the promoters relative to the transcription start or end site (green).
 Click OK.
 |
Next you need one or multiple motifs. Jaspar and the public part of Transfac are by default in MotifLab but you can add your own or other public motif collections.
| How to import motifs ? |
|---|
| In the Motifs section of the left menu click the +sign and select MotifCollection.
 As stated previously MotifLab has a number of predefined motif collections. Choose Jaspar Core and click OK.
 MotifLab has now loaded all Jaspar core motifs as you see in the Motifs section of the left menu. We only want to use motifs from human. To do this
 MotifLab creates a new motif collection called MotifCollection2 with only the 79 human motifs. Deselect Jaspar Core, deselect MotifCollection 2 and select it again. Check if all motifs from MotifCollection2 are selected.
 |
| How to scan the sequences for potential matches to the motifs ? |
|---|
 |
DNA sequences contain repeat regions that might interfere with the motif searches. It is recommended to remove repeat regions from your sequences. MotifLab allows you to do so.
Motif searches always generate an enormous amount of false positives (regions in the DNA sequence that are similar to the motif but do not actually bind to the TF). You can reduce the number of false positives by focussing on regions in the promoter that are conserved between species. This is called phylogenetic footprinting. It is assumed that regions that are functionally important (like TFBS) do not change during evolution because if they change, their function would be lost.
MotifLab allows you to add feature annotation tracks to display and use annotation of the sequences, like the presence of conserved regions, repeats, open chromatin regions...
We will use the following tracks: Conservation and RepeatMasker
| How to add these feature annotation tracks? |
|---|
| Click the Add Feature Data Track button in the top menu
 This opens the Datatracks dialog. Here you can select the tracks you want to visualize. Tracks with red buttons are not available for the organism you are working on.
 The annotation is now visualized. |
Now you can use the annotation to remove motifs that are located in repeat regions or in regions with low conservation.
| How to filter motifs in repeat regions or regions with low conservation? |
|---|
| Right click the binding sites track and select Perform Operation -> Transform -> Filter
 This opens the Filter dialog. Set the condition used for the filtering as follows
 This will remove motifs in regions with low conservation.
 Set the second condition as follows
 Right the AND and select Change Operator to OR to remove motifs that are in low conserved regions OR that are located in repeats.
 You see that this greatly reduces the number of motifs that are shown in the track. The only motifs that are retained are those in conserved regions and outside repeat regions.
 |
You can do the same with other annotations e.g. remove motifs in closed chromatin regions...
You should, however, when you do this for real, use a better algorithm than the Simple Scanner. You can for instance download and add Clover.
Finding enriched TF binding motifs in a list of genes via Pscan
We performed additional experiments to find additional genes that show the same expression pattern as the ones in the previous exercise, so we now have an extended list of coexpressed genes.
| Using PScan can you find other TF binding motifs that are enriched in the promoters of our coexpressed genes ? |
|---|
Pscan accepts the following gene or transcript identifiers: RefSeq (for human, mouse, and drosophila, e.g. NM_000546) TAIR (e.g. AT1G08810) for Arabidopsis; SGD (e.g. YPL248C) for yeast.

The output shows the ranking of the TFs selected according to the enrichment p-values of their PWM.
 You see that the TP53 motif is strongly enriched in the promoters of these genes. Additionally you also see enrichment of motifs that bind HFN1A, a tumor marker just like TP53. By clicking on a matrix name, you can open a dedicated page showing detailed results: matrix, logo, and links to the database entry as well as to the ID (PMID) of the PubMed entry describing the generation of the PWM.
 A simple graphic representation shows the average matching value of the matrix in the sequences in our list of TP53 targets (top) compared to the average matching value and standard deviation (in green) on a set of all promoters (same size of regions with respect to the TSS as selected) of the same organism (bottom).
 Sample statistics shows the statistics of the enrichment analysis: the p-value, the Bonferroni corrected p-value, mean and standard deviation of the matching score on the list of TP53 targets, and number of sequences in the list. These latter pieces of information can be useful to compare the results of different data sets by using the Compare with.. box next to the statistics. |
The p-value should be read as: "If we take as many random genes from the same organism as in the input set, and we select their corresponding promoter regions, what is the probability of having the same score as obtained in the input set?"
However, Pscan checks 130 matrices (or 280 if you select Transfac) so 130 independent statistical tests are performed. This means you have to correct for this if you do not want to end up with a long list of false positives (the p-value implies that the TFBS is enriched while in fact it is not).
Pscan uses a Bonferroni correction, meaning that the typical significance threshold (0.05 or 0.01) is divided by the number of tests (in our case: 130). This means that a significance threshold of 0.05 becomes 0.0003 after Bonferroni correction. When you use this value as significance threshold, the score for HFN1a is not significant. However, it should be noted that if you do so many t-tests the Bonferroni correction is not the ideal choice: it's too stringent in these cases.
In the exercises above we searched for known TF binding motifs but often you do not know the motif that regulates the expression of your genes of interest and you have to search for unknown motifs.
There are several tools to solve the complex problem of de novo identification of motifs that are enriched in a list of sequences. The wikipedia page has a very nice overview on these tools (see [1]).
What you need as an input is a list of promoters of which you are more or less certain that they all bind to the same TF, you just don't know which TF. If you have such promoter sequences you can search for common motifs that occur in all promoters.
Besides the very popular MEME suite, you can also use one of the RSAT tools for this.
You have to do the following steps to do the analysis:
- Get promoter sequences using the RSAT tool retrieve Ensembl seq or retrieve sequence
- Analyse them for common motifs using one of the RSAT tools under Motif discovery
EMBOSS: translation of nucleic acid into protein
The majority of protein sequences are derived from annotated nucleic acid sequences using a translation tool.
- Go to the EMBOSS home page
- Search the page for translation (using Ctrl + F)
- In the NUCLEIC TRANSLATION section in the left menu, click transeq

| Translate the CDS of the mouse basic domain/leucine zipper transcription factor L36435. |
|---|
If you did everything right you should obtain a protein sequence starting with a methionine (M) and ending with a *, which represents a stop codon. |
| Translate the CDS of the unknown human sequence that you can download here. |
|---|
| In this sequence you don't know where the CDS is located but you do know it's there. Fortunately you can use transeq to find a CDS without knowing in which frame you should look. To do this you can select 6 (All six fames) in the Parameters section of the tool to obtain all reading frames.
 |
Of course, this doesn't work when you start from a genomic sequence of a gene that contains introns. Translation tools do not take into account the presence of introns.
A good alternative for the EMBOSS Transeq tool is the Expasy Translate tool.
GENSCAN to predict eukaryotic genes
Translating DNA (see previous exercise) and gene prediction are two very different things.
The translation software will translate any DNA sequence that you feed it regardless of whether the DNA sequence really is a CDS or not. Gene prediction software on the other hand looks for sound evidence (base composition, similarity to known CDS, presence of motifs...) to predict the location of CDS in a DNA sequence.
The Genbank entry with accession number X02419 contains the sequence of the gene encoding the urokinase-type plasminogen activator. We will run gene prediction software on the sequence and see if the software manages to correctly find the CDS. GENSCAN is one of the most popular tools for gene searching. It is used by the genome annotation pipeline at EBI, while a slightly modified version (Gnomon) is used at NCBI.
GENSCAN models eukaryotic genes with HMMs (hidden markov models) for the:
- coding parts of eukaryotic exons
- TATA-box
- CAP-site
- polyA-site
The software offers these models for human (usable for all vertebrates), Arabidopsis thaliana and maize.
GENSCAN is available via the Mobyle portal. Go to the Mobyle home page.
- In the left menu expand sequence
- Expand nucleic
- Expand gene_finding
- Click genscan

In the Input section:
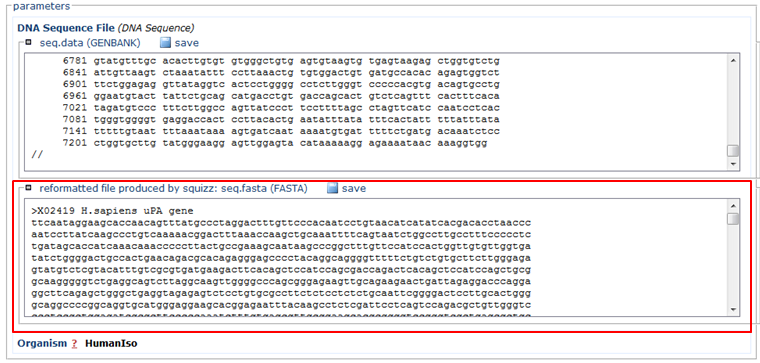
- Paste the sequence in FASTA format (red)
- In the Organism section, choose HumanIso (green)
- Click the Run button (blue)

The Mobyle server will ask for your email address, provide it and click Ok.
The Mobyle server will ask you to validate your submission, do so and click Ok.
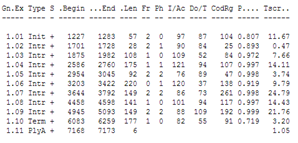
You can compare the result with the annotation in the Genbank file of the sequence: it has Genbank accession X02419.
| Did GENSCAN predict the CDS correctly ? |
|---|
| You can choose to see the GENSCAN prediction full screen The Mobyle portal is not always working well so if it doesn't generate an output you can go to the genscan page at MIT. Copy the reformatted sequence from the Mobyle results page: and paste it in the GENSCAN page at MIT in the input sequence text box. Click Run Genscan. You get the same results as shown above for the Mobyle portal. The annotation of the Genbank file: You will see that GENSCAN correctly predicted the CDS. |
GENSCAN also found a polyA-site in the correct location. Note that GENSCAN does not find non-coding exons like the first exon in the Genbank file.
Selecting primers for PCR
Designing regular PCR primers using Primer3
The RefSeq entry NM_079400 contains the sequence of the D. melanogaster mRNA coding for tap, the target of Poxn. Tap encodes a bHLH protein expressed in larval chemosensory organs and involved in the response to sugar and salt. We wish to amplify by PCR the region coding for the HLH motif.
| Find out which part of the sequence corresponds to the Helix-loop-helix domain. |
|---|
|
Go to the Genbank record and click on the Gene link or go directly to the Gene record via GQuery.
 In the NCBI Reference Sequences section of the Gene record you can find the location of the domain: 155-211.
 This location applies to the protein sequence so it's expressed in amino acids. This means you have to calculate the corresponding positions in the nucleotide sequence:
In the nucleotide sequence of the RefSeq mRNA record, the domain is located between position +577 and +745. |
The Primer3 program can help to select suitable primers.
Go to Primer3 [1] or use the nicer interface at Primer3Plus [2] (in the solution below Primer3Plus was used).
| Use Primer3 to select primers to amplify the HLH motif of tap. |
|---|

This is, however, not what we want: we want to amplify the hlh region only so we aim for a much smaller product. Since the hlh region is 168 bp long we set this as the lower limit for the product size range and we'll take 300 bp as the upper limit.
 Click Pick Primers
 |
In the results page you can see the following:
The program has selected 5 primer pairs, with the best at the top.
For each primer you can find:
- the location on the sequence where the first base of the primer binds (for the forward/left primer this is on the complementary strand)
- its length
- its melting temperature
- its %GC
- the score of an alignment of the primer with the other primer and with itself
The program first considers all possible primers and retains those that satisfy a series of criteria, then considers all possible pairs of these primers and retains those who satisfy a series of criteria. At the bottom of the page you can see information about how many primers were considered and rejected/retained.
| Check the specificity of the first set of primers. |
|---|
|
Click the Send to Primer3Manager button (red) for the first primer pair.
 This leads to a form for further evaluating the primers:
 Click the BLAST! link to check the specificity of the primers.
 |
Look at the 8 highest scoring hits for the forward primer.
| Is the forward primer really identical to 8 different locations of the Drosophila genome ? |
|---|
|
 No what you see here are 8 records that all contain the tap sequence (mRNA, chromosome, synthetic constructs...) Remarkably some of them are annotated as the biparous TF, but if you take a closer look you will see that tap and biparous are the same gene. This is the problem when working with the nr database which in contrast to whats it's name lets you believe is not non-redundant at all. |
So it's always a better idea to Blast against Refseq sequences unless they are not available yet for the organism you work with or you have reasons to believe that they are not complete (i.e. they do not represent the full genome). However, for model organisms, full chromosome sequences are available so you can use Refseq for BLAST.
| Repeat the Blast on RefSeq |
|---|
Now you see that the redundancy has disappeared. |
Go to the results of the reverse primer.

When evaluating the specificity of the primers, it is especially important to check that the primers are specific at the 3' end because that's the site where the polymerase will attach nucleotides.
So generally, it is recommended to not use primers that contain long identical stretches (>15 nt for primers of 20 nt long) to other regions in the genome apart from the one you want to amplify/measure, and certainly not if these stretches comprise the last nucleotide at the 3' end of the primer.
When you look at the alignment of the first hit on chromosome X of the reverse primer you see that this is a borderline case: an identical stretch of 15nt including the nucleotide at the 3' end !

Designing regular PCR primers using PrimerBLAST
It's a lot of work to design primers using Primer3/CLC Main Workbench especially if you discover during the BLAST that the suggested primers are not specific and you have to start all over again.
Therefore, we will now design primers using the PrimerBLAST tool [3] of NCBI. PrimerBLAST uses the Primer3 algorithm to find primers allowing you to set all the parameters related to effciency of the primers (e.g. secondary structure and dimer formation, Tm, difference in Tm...). On top of this, PrimerBLAST uses the BLAST algorithm to find specific primers, that will only bind to the intended target. It is the only tool that incorporates the BLAST in the primer search.
For some applications, it is vital to have specific primers. This is for instance the case when you use PCR for quantification (qPCR) or when you want to amplify a commonly occurring sequence from the genome.
As an example, we will design primers to amplify a region of RefSeq entry NM_079400, which represents the D. melanogaster mRNA coding for tap, the target of Poxn. Tap encodes a bHLH protein expressed in larval chemosensory organs and involved in the response to sugar and salt. We wish to amplify the region encoding the Helix-loop-helix domain by PCR. In the nucleotide sequence of the RefSeq record, the domain is located between position +577 and +745.
So, let's go to the PrimerBLAST page.
| How to set the general primer parameters ? |
|---|
| Fill in the first two section of the BLAST form as follows:
 Note that you do not have to fetch the sequence, you can directly enter the RefSeq ID (red). To amplify the helix-loop-helix domain you need a primer at both sides of the domain. To accomplish this you need to define the regions in the sequence that you want to use for selecting primers in by defining a range for both primers as shown in green. Set the product size (blue) and the maximum difference in anneal temperatures of the primers of a pair (purple) to reasonable values (e.g. take the same values as in Primer3). |
Remember that PrimerBLAST also allows you to set the parameters related to specificity of the primers.
| How to set the parameters related to primer specificity ? |
|---|
| Fill in the first two section of the BLAST form as follows:
 Compare the primers to the Drosophila (green) genome (red). You see that you can easily define the specifity that you want to obtain (blue): you can set
|
Only primers that fulfill the specificity criteria that you have set will be shown.
| How to set the parameters related to primer efficiency (hairpins, dimers, length...) ? |
|---|
| Expand the Advanced parameters section:
 This shows a set of advanced parameters concerning the primer specificity (Primer Pair Specificity Checking Parameters). I normally keep these at their default settings. If you want to make your search more stringent you can decrease the Blast word size. This is the minimum number of consecutive nucleotides that have to exactly match between the primer and a sequence from the database to consider it a hit. The lower you set it, the faster BLAST will consider a sequence from the database a hit. After that follow the advanced Primer parameters. These are the ones that we are interested in. At the top you have a set of general parameters that are quite self-explanatory.  Then follow the parameters concerning the likelihood that the primers will form hairpins and dimers. By default Primer-BLAST uses secondary structure alignment to predict these characteristics but you can choose to use more advanced and accurate thermodynamic models. Calculations will be slow when you choose these models though. Basically, instead of looking at individual nucleotide matches you use the stability of dinucleotide matches, e.g. the stability of CT hybridized to GA is different from that of CA hybridized to GT. The scores for self and pair complementarity in the default secondary structure are local alignment scores: the scoring system gives 1.00 for complementary bases, -1.00 for a mismatch, and -2.00 for a gap. So a score of 8 corresponds to 8 consecutive matches or 9 aligned nucleotides with 8 matches and 1 mismatch...  |
Click the Get Primers button (purple) to start the search. After a few seconds the search returns 5 specific high quality primer pairs:

Knowing that PrimerBLAST uses the same algorithm to pick primers as Primer3 [4], PrimerBLAST will save you a lot of work and time when you have to design primers.
Designing primers for qPCR
Exercises on qPCR primer design can be found on our primer design wiki page.
References: